Figure 1.
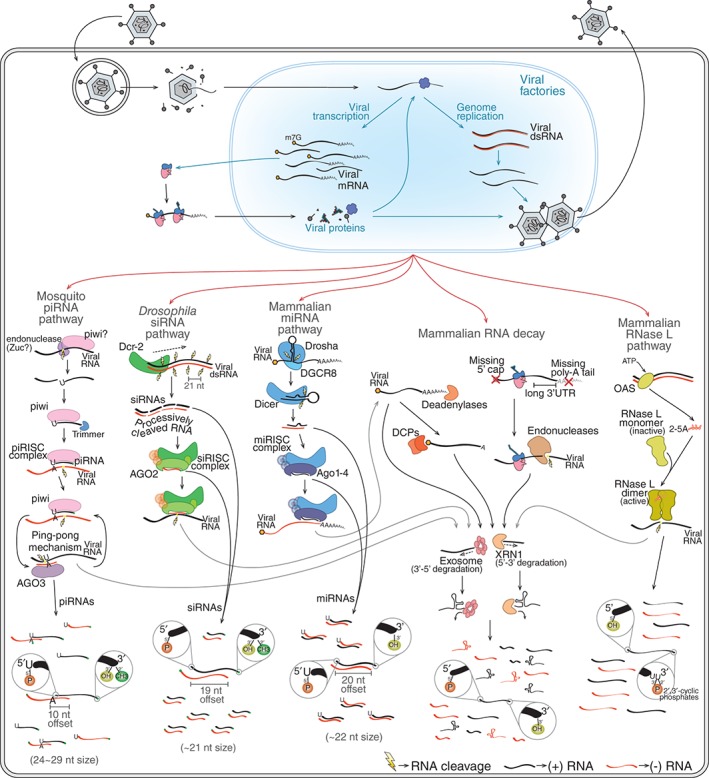
The generation of virus‐derived small RNAs by host pathways. During infection, virus genomes and transcripts are sometimes exposed and can be recognized by different host RNA surveillance mechanisms. RNA decay, RNA interference (RNAi), and RNase L pathways are good examples of such mechanisms. Mammalian RNA decay involves different mechanisms such as deadenylation‐dependent and nonsense‐mediated decay (NMD). The former involves shortening of the poly(A) tail by deadenylases followed by removal of the 5′ cap by mRNA‐decapping enzymes (DCPs). NMD is initiated by recognition of aberrant mRNAs that are cleaved by endonucleases. In both cases, the initial cleavage allows the mRNA to be targeted 5′→3′ by XRN1 and 3′→5′ by the exosome complex. The mammalian RNase L pathway is triggered when viral dsRNA is recognized by OAS enzymes that catalyze the production of 2′‐5′ oligoadenylates (2‐5A). These molecules induce dimerization and activation of RNase L that cleaves single‐stranded RNAs mostly at U‐rich regions. Different RNAi mechanisms can generate vsRNAs during viral infection including the mammalian miRNA pathway, Drosophila small interfering RNA (siRNA) pathway, and mosquito piRNA pathway. The Drosophila siRNA pathway is activated by Dcr‐2‐mediated recognition of viral dsRNA that is processes progressively to generate phased duplex siRNAs. siRNA duplexes are loaded onto AGO2 to generate siRISC that will find and cleave complementary RNAs. The mammalian miRNA pathway is initiated by the recognition of structure regions within long transcripts in the nucleus by the RNase III Drosha. This enzyme, in association with a partner protein known as DGCR8, cleaves the primary transcript to excise a short hairpin (~65 nt) that is then exported to the cytoplasm. There, the hairpin is further processed by Dicer to generate miRNA duplexes of ~22 nt that will be loaded onto different mammalian Argonaute proteins (Ago1–4) to form miRISC. This complex targets complementary regions within the 3′ UTR of mRNAs leading to translation inhibition. The mosquito piRNA pathway is triggered by the recognition of single‐stranded RNA precursors in a manner dependent on a specialized group of Argonautes known as PIWI proteins usually associated with an endonuclease known as Zucchini (Zuc). This initial recognition triggers processing of the precursor into primary piRNAs that remain associated with PIWI proteins to form the piRISC. This complex carries out cleavage of complementary RNA and can also initiate the production of more piRNAs. These secondary piRNAs require an amplification loop, referred to as the ping‐pong mechanism, that involves another Argonaute protein known as AGO3. Different RNA surveillance mechanisms may work together to generate vsRNAs. For example, RNA fragments generated by RNAi and RNase L pathways can be further targeted by RNA decay mechanisms. RNA surveillance mechanisms generate vsRNAs that have unique molecular characteristics such as terminal modifications, size, strand bias, and nucleotide preferences shown in the figure.
