Figure 3.
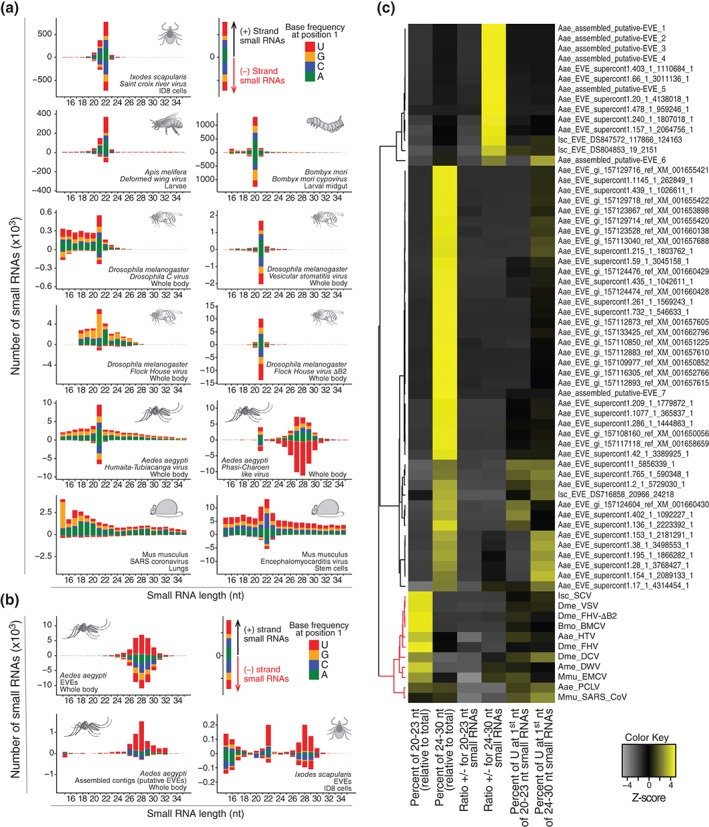
Activation of the small interfering RNA (siRNA) pathway is a common and specific response to virus infection. (a) Virus‐derived small RNAs in different animals often show a profile consistent with the activation of the siRNA pathway. Virus‐derived siRNAs range from ~20 to 23 nt and are symmetrical in polarity and base preferences. (b) Endogenous viral elements (EVEs) represent remnants of virus sequences integrated into animal genomes and also generate small RNAs. EVE‐derived small RNAs are often ~24–30 nt, asymmetrical in polarity, and base preferences that is consistent with a piRNA signature. (c) Size, polarity, and nucleotide preferences were determined for small RNAs derived from EVEs and active viruses. These molecular footprints were then used to perform hierarchical clustering of different EVEs and viruses. The small RNA pattern clearly separates clusters containing viruses (in red) and EVEs (in black). Notably, within the cluster of viruses, we observe two small subclusters. Virus grouped in the larger subcluster show a classical siRNA signature even when the pathway is partially inhibited such as for FHV and DCV in Drosophila. The second subcluster contains the vsRNA profile of coronavirus in mouse lungs and PCLV in mosquitoes, both of which show a more divergent profile from the siRNA signature. The data in this figure were obtained from the analysis of small RNA libraries from published studies (accession numbers: SRR1803378 , SRR1803382 , ERR555100, ERR274423, ERR654010, SRR1803383 , GSM792688, GSM792692, SRR452408, and SRR640612).10, 12, 77, 78, 79, 80, 81, 82
