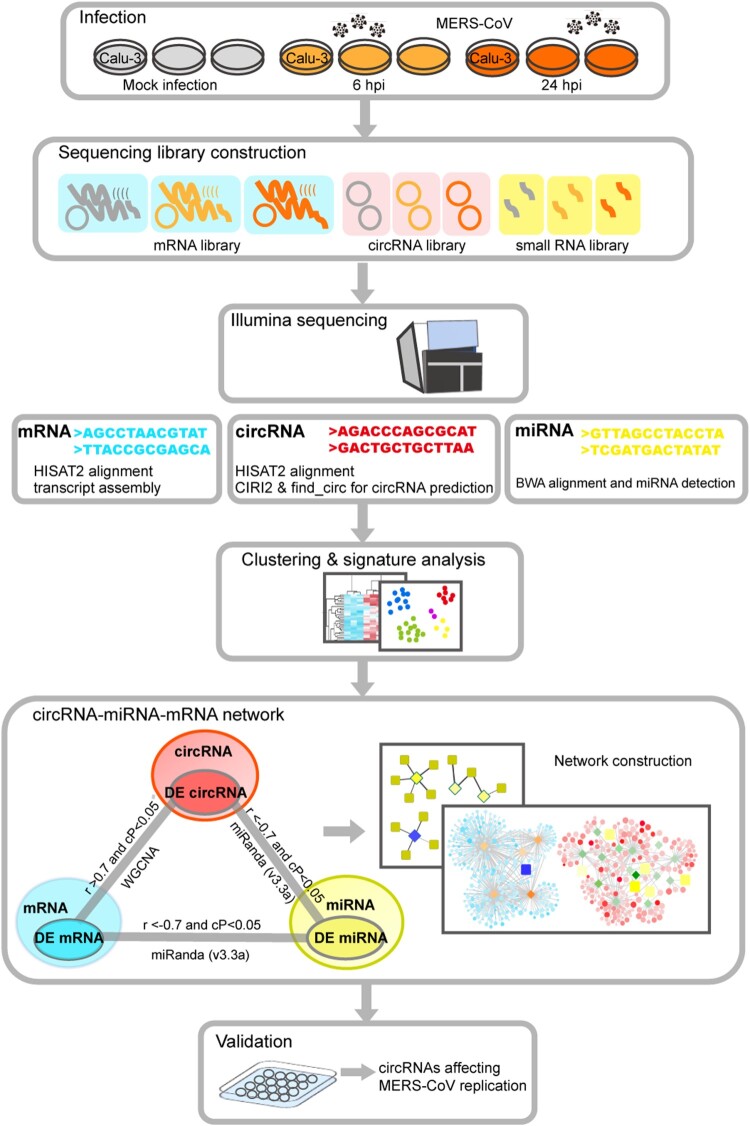
Figure 1.
Schematic overview of RNA sequencing, data analysis, and critical pathogenic circRNAs identification. Mock-infected and MERS-CoV-infected Calu-3 cells were harvested for RNA sequencing. CIRI2 and find_circ were used for circRNA prediction. Differential expression analysis and co-expression analysis were implemented to profile the impact of MERS-CoV infection on host cells and construct the circRNA-miRNA-mRNA co-regulatory network. The effects of selected circRNAs identified in the circRNA-miRNA-mRNA network on MERS-CoV replication were validated in vitro.