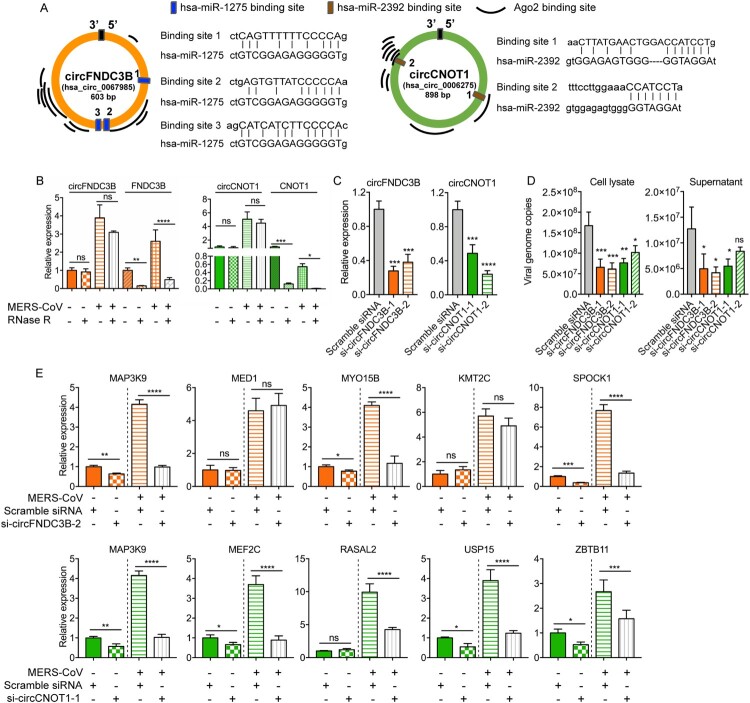
Figure 7.
Inhibitory effect of circFNDC3B and circCNOT1 knockdown on MERS-CoV replication. (A) The Ago2 protein and miRNA binding sites of circFNDC3B and circCNOT1 predicted by CircInteractome and miRanda, respectively. (B) qPCR validating the expression and RNase R resistance property of the circRNAs circFNDC3B and circCNOT1, and the mRNAs FNDC3B and CNOT1. (C) qRT-PCR examining the knockdown effect of siRNA candidates. Linear RNA of GAPDH was used as internal reference for normalization. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001; ****P-value < 0.0001, one-way ANOVA. (D) Depletion of circFNDC3B and circCNOT1 suppressed MERS-CoV replication in cell lysate and supernatant. Scramble siRNA was served as a negative control. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001; ****P-value < 0.0001, one-way ANOVA. (E) CircFNDC3B and circCNOT1 knockdown decreased the expression of representative targeting genes. Student’s t-test was adopted to calculate the significance of gene expression with or without MERS-CoV infection. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001; ****P-value < 0.0001.