Abstract
Extracytoplasmic sugar decoration of glycopolymer components of the bacterial cell wall contributes to their structural diversity. Typically, the molecular mechanism that underpins such a decoration process involves a three-component glycosylation system (TGS) represented by an undecaprenyl-phosphate (Und-P) sugar-activating glycosyltransferase (Und-P GT), a flippase, and a polytopic glycosyltransferase (PolM GT) dedicated to attaching sugar residues to a specific glycopolymer. Here, using bioinformatic analyses, CRISPR-assisted recombineering, structural analysis of cell wall–associated polysaccharides (CWPS) through MALDI-TOF MS and methylation analysis, we report on three such systems in the bacterium Lactococcus lactis. On the basis of sequence similarities, we first identified three gene pairs, csdAB, csdCD, and csdEF, each encoding an Und-P GT and a PolM GT, as potential TGS component candidates. Our experimental results show that csdAB and csdCD are involved in Glc side-chain addition on the CWPS components rhamnan and polysaccharide pellicle (PSP), respectively, whereas csdEF plays a role in galactosylation of lipoteichoic acid (LTA). We also identified a potential flippase encoded in the L. lactis genome (llnz_02975, cflA) and confirmed that it participates in the glycosylation of the three cell wall glycopolymers rhamnan, PSP, and LTA, thus indicating that its function is shared by the three TGSs. Finally, we observed that glucosylation of both rhamnan and PSP can increase resistance to bacteriophage predation and that LTA galactosylation alters L. lactis resistance to bacteriocin.
Keywords: cell wall, glycosylation, glycosyltransferase, bacteriophage, glycobiology, genomics, peptidoglycan, flippase, glycopolymer, lactic acid bacteria, lipoteichoic acid (LTA), phage receptor, glycopolymer, flippase
Introduction
The cell wall of Gram-positive bacteria plays a protective role in maintaining cell integrity, yet also makes the bacterial cell vulnerable because it mediates interactions with phage predators or host immune defenses. Substantial knowledge has been accumulated regarding the molecular and biochemical steps involved in the assembly of various cell wall constituents, including the major one, peptidoglycan (1), as well as other constituting glycopolymers, such as lipoteichoic acids (LTAs)5 (2), wall teichoic acids (WTA) (3), and cell wall polysaccharides (CWPS) (4). Bacterial glycopolymers are characterized by their astonishing structural diversity. A three-component glycosylation system (TGS) has previously been proposed for the extracytoplasmic addition of sugar residues on bacterial glycoconjugates, such as teichoic acids (TAs) in Gram-positive bacteria, thus contributing to generate structural diversity (5). A TGS is characterized by the initial synthesis of an undecaprenyl-phosphate (Und-P)-sugar intermediate, catalyzed by a membrane-anchored glycosyltransferase (GT) at the inner face of the cytoplasmic membrane. Following this, a so-called flippase reorients the above-mentioned membrane-associated sugar intermediate from the cytoplasmic to the periplasmic side (or outer side) of the membrane. The final attachment of the sugar onto the glycoconjugate is catalyzed by an integral membrane GT, which contains between eight and 13 transmembrane helices (TMHs) and possesses a GT-C fold (5). In particular, the three functions involved in this process have been attributed to specific genes in Listeria monocytogenes (6, 7): GtcA (a flippase enzyme), GtcB (Und-P GT), and GtcC (a polytopic transmembrane (PolM) GT) were shown to be involved in the galactosylation of WTA in L. monocytogenes serotype 4nonb (7). Additionally, GtlA (Und-P GT) and GtlB (PolM GT) were shown to attach galactose (Gal) onto the LTA moiety of L. monocytogenes 10403S (6).
Various Lactococcus lactis strains produce an apparently unique dual-component CWPS structure (8): a rhamnose-rich and unexposed (i.e. peptidoglycan-embedded) polysaccharide chain, known as the rhamnan (9), and a surface-exposed phosphopolysaccharide or so-called polysaccharide pellicle (PSP) (10, 11). The biosynthetic machinery responsible for the production of these two components is encoded by a large (25–30-kb) gene cluster (the cwps gene cluster) (10). Through the application of transposon-mediated, spontaneous, or directed mutagenesis strategies, the importance of the PSP for bacteriophage infection has been defined (10–13). The PSP subunits exhibit a certain degree of structural diversity between L. lactis strains. For example, the PSP of both L. lactis MG1363 and L. lactis SMQ-388 is a polymer of phosphohexasaccharide subunits containing a Glc side chain (11, 14), whereas the PSP of L. lactis 3107 is composed of phosphopentasaccharide linear repeating units (12). Interestingly, the PSP subunit structures of L. lactis SMQ-388 and L. lactis 3107 are virtually identical with the exception of the presence of the Glc side chain in the structure of the former. The genetic divergence of the cwps gene cluster of the two strains does not appear to account for the emergence of such a structural discrepancy in their PSP, prompting an investigation into the existence of a potential TGS in L. lactis similar to those discovered in L. monocytogenes.
In the current study, we demonstrate the existence of three sets of gene pairs in L. lactis NZ9000 (llnz_00690 and llnz_00695, here termed csdA and csdB; llnz_03080 and llnz_03075, here termed csdC and csdD; and llnz_07820 and llnz_07825, here termed csdE and csdF), whose products exhibit sequence and topological identity to the two GTs of the previously characterized L. monocytogenes TGS (6, 7). The function of each gene set was characterized in L. lactis NZ9000, in which they were shown to be required for the glycosylation of a distinct glycopolymer within the cell envelope. CsdAB was shown to glucosylate the rhamnan, CsdCD was shown to glucosylate the PSP, and, finally, CsdEF is involved in LTA galactosylation. Additionally, a candidate flippase-encoding gene (llnz_02975, here renamed cflA), was identified, and its functionality was characterized. Our results show that its product, CflA, forms part of the three glycosylation pathways, thus completing the putative TGSs. Finally, the effect on fitness and bacteriophage resistance conferred by the sugar substitutions of the above-mentioned glycopolymers is reported.
Results
Identifying lactococcal homologues of L. monocytogenes gtcABC and gtlAB
Previous NMR structural analysis of the CWPS in certain L. lactis strains identified the presence of a Glc side chain on the PSP subunit produced by L. lactis MG1363 (11) and L. lactis SMQ-388 (14). This feature is absent in the PSP subunit of L. lactis 3107, which was unexpected given the high sequence identity between the 3′-region of the cwps gene clusters of strains 3107 and SMQ-388 (12), whereas all three strains also harbor the same number of putative glycosyltransferase-encoding genes in their respective cwps clusters. This observation suggests that additional genes located outside the cwps gene cluster contribute to the biosynthesis of the final PSP structure of certain strains.
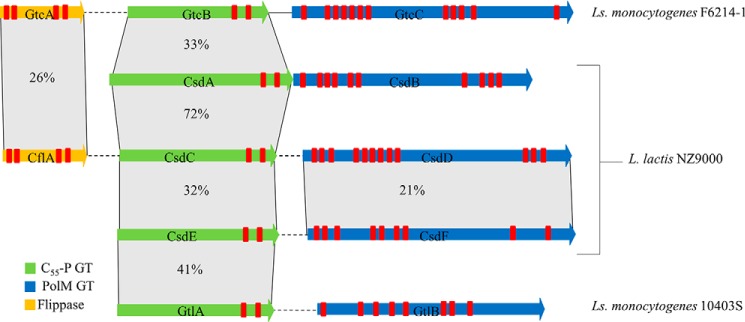
Recently, genes had been identified in L. monocytogenes that are responsible for the attachment of sugar side chains on LTA (i.e. gtlA and gtlB) (6), or on WTA (i.e. gtcA, gtcB, and gtcC) (7). We used the corresponding protein sequences to identify genes that may be responsible for the Glc substitution of the PSP of L. lactis NZ9000. The L. lactis NZ9000 genome contains three gene pairs whose products exhibit sequence similarity to and share transmembrane topology properties with the L. monocytogenes proteins: the putative Und-P GT–encoding genes llnz_00690 (csdA), llnz_03080 (csdC), and llnz_07820 (csdE), as well as their adjacent and putative PolM GT-encoding genes llnz_00695 (csdB), llnz_03075 (csdD), and llnz_07825 (csdF), respectively (Fig. 1). At protein level, the three predicted Und-P GTs encoded by L. lactis NZ9000 exhibit varying levels of similarity to each other and to the L. monocytogenes GtcB (33–72% amino acid identity), whereas the putative PolM GT proteins do not exhibit such a high degree of identity (Fig. 1). CsdA, CsdC, and CsdE belong to the GT2 family (CAZY; RRID:SCR_012909) that contains enzymes transferring sugar from NDP-sugar to various substrates, including Und-P. They contain two TMHs at their C terminus and the typical DXD conserved motif (92DVD94 in CsdA, 95DAD97 in CsdC, and 102DAD104 in CsdE) of GT-A fold enzymes, which interacts with the phosphate group of nucleotide donors. The three proteins CsdB, CsdD, and CsdF contain 10, 12, and 9 predicted TMHs (TMHMM analysis), respectively. In addition, HHpred analysis predicts structural similarity of these three proteins with ArnT, a typical GT with a GT-C-fold that is found in modifying GTs of TGSs. Interestingly, a gene encoding a protein with sequence similarity to the third protein involved in the glycosylation of L. monocytogenes WTA (GtcA), which is presumed to encode a flippase, is present in the L. lactis NZ9000 genome (llnz_02975, designated here as cflA (carbohydrate flippase)). Similar to GtcA, CflA is a predicted small (148 amino acids) integral membrane protein containing a total of four TMHs, as found previously in polyprenyl-monophosphate sugar flippase candidates of TGSs (7).
Figure 1.
Schematic representation of a locus map showing three of the genes, gtcABC, associated with WTA galactosylation in L. monocytogenes F6214-1 and two genes, gtlAB, associated with LTA galactosylation in L. monocytogenes 10403S along with seven genes identified in L. lactis NZ9000 by sequence homology search. Genes are color-coded according to predicted functional domains: green, Und-P GT; blue, PolM GT; yellow, flippase. Percentage similarity based on BLASTP alignment (of the encoded protein products) is also indicated. Finally, sequences representing the predicted TMHs (predicted with TMHMM server version 2.0) are indicated by red rectangular bands. Solid lines, adjoining genes; dashed lines, order/orientation of genes altered for visualization purposes.
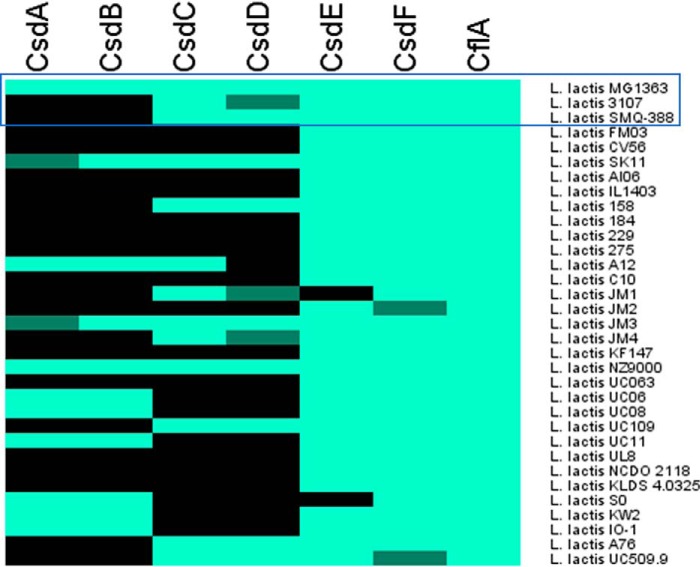
The presence of the three identified L. lactis NZ9000 gene pairs within the genomes of L. lactis 3107 and SMQ-388 was investigated through sequence alignments (Fig. 2), revealing that only csdEF is shared by all three strains. L. lactis SMQ-388 also contains homologues of csdCD, but appears to lack csdAB, whereas L. lactis 3107 lacks both csdAB and a functional copy of csdCD, due to a transposase insertion within csdD.
Figure 2.
Heat map representing the presence (light green), absence (black), or nonfunctionality (dark green) of each of the six proteins encoded by the three glycosyltransferase-associated gene pairs and the flippase (CflA) in a total of 32 L. lactis publicly available genomes as well as L. lactis SMQ-388. The heat map was generated in Mev (V4_9_0). The top three strains, for which PSP structure was previously established by NMR, are highlighted by a blue frame.
Prevalence of the genes encoding putative sugar decoration systems
A genome-wide search of 32 publicly available genomes of L. lactis, in addition to L. lactis SMQ-388, failed to identify any additional candidate gene pairs that may encode components of TGSs other than those presented in Fig. 1. Sequence comparisons at protein level indicated the almost ubiquitous presence of csdEF in L. lactis genomes (29 of 33 strains), whereas csdAB and csdCD are much less widely distributed (9 and 10 strains of a total of 33 examined strains, respectively) (Fig. 2). A small subset of the genomes appears to contain a functional copy of one of the two genes in a given gene pair, whereas the other is predicted to be nonfunctional due to sequence frameshifts or inserted transposon elements. Furthermore, a protein of high sequence similarity to CflA, with a predicted flippase funtionality, is encoded by all assessed strains. Interestingly, the genomic region surrounding csdCD is either immediately flanked or interrupted by transposase-encoding genes, or it forms part of a predicted prophage region. Based on these results and in association with the structural data regarding the glucose side chain found in the PSP of both L. lactis MG1363 and L. lactis SMQ-388 (and its absence in L. lactis 3107), the csdCD gene pair appeared to be the most likely candidate responsible for the attachment of the glucose side chain to PSP.
CsdCD is involved in PSP side-chain glycosylation
To ascertain whether the enzymes encoded by csdCD contribute to the final PSP structure, several mutants/derivatives of L. lactis NZ9000 and 3107 were generated. L. lactis NZ9000 derivatives carrying nonsense mutations in either csdC or csdD or both were generated (Table 1). In parallel, the csdCD gene pair was expressed in L. lactis strain 3107, which lacks functional csdCD homologs (Table 1).
Table 1.
Strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Feature(s) | Source |
|---|---|---|
| Bacterial strains | ||
| L. lactis subsp. cremoris NZ9000 | L. lactis MG1363 derivative containing nisRK, host to phages jj50, p2, and sk1 | Ref. 46 |
| L. lactis subsp. cremoris 3107 | Host to phages LC3, TP901–1, 66901, 66902, 66903, 62604, 62605, and 62601 | Ref. 47 |
| L. lactis subsp. cremoris VES5751 | L. lactis MG1363 derivative exhibiting a deficient PSP phenotype due to a mutation in llmg_0226 | Ref. 11 |
| L. lactis subsp. cremoris NZ9000-csdAB | NZ9000 with GAATTCG insert in llnz_00690 (csdA) and llnz_00695 (csdB), resulting in a TGA stop codon insertion in each gene | This work |
| L. lactis subsp. cremoris NZ9000-csdC | NZ9000 with GAATTCG insert in llnz_03080 (csdC) resulting in a TGA stop codon in csdC | This work |
| L. lactis subsp. cremoris NZ9000-csdD | NZ9000 with GAATTCG insert in llnz_03075 (csdD) resulting in a TGA stop codon in csdC | This work |
| L. lactis subsp. cremoris NZ9000-csdCD | NZ9000 with GAATTCG insert in llnz_03075 (csdD) and llnz_03080 (csdC) resulting in a TGA stop codon insertion in both genes | This work |
| L. lactis subsp. cremoris NZ9000-csdEF | NZ9000 with GATAACCC insert in llnz_07820 (csdE) and llnz_07825 (csdF), resulting in a TGA and TAA stop codon insertion, respectively | This work |
| L. lactis subsp. cremoris NZ9000-cflA | NZ9000 with TAATAGGGG insert in llnz_02975 (cflA) resulting in a TAA and TAG double stop codon in cflA | This work |
| Plasmids | ||
| pJP005 | Recombineering-facilitating vector containing recT, PnisA, Cmr | Ref. 39 |
| pNZ44 | High-copy expression vector, contains P44 constitutive promotor, Cmr | Ref. 37 |
| pNZ44str | Derivative vector of pNZ44, Strr | Developed by Andrea Garzon, UCC Collection |
| pCNR | Recombineering-facilitating vector containing recT, PnisA, Cmr derived from the low-copy vector pJP005 | This work |
| pVPL3004 | Low-copy vector expressing cas9 along with tracRNA, Eryr | Ref. 41 |
| pCRISPR | High-copy vector carrying CRISPR repeats and used for inserting target spacer sequences, Tetr | Ref. 41 |
| pCRISPR::cflA | pCRISPR plasmid carrying a CRISPR repeat targeting the recombineered sequence of gene cflA, Tetr | This work |
| pNZ44::csdCD | pNZ44 containing genes csdC and csdD | This work |
| pNZ44::csdA | pNZ44 containing gene csdA | This work |
| pNZ44::csdB | pNZ44 containing gene csdB | This work |
| pNZ44::csdAB | pNZ44 containing genes csdA and csdB | This work |
| pNZ44str::csdAB | pNZ44str containing genes csdA and csdB | This work |
| pNZ44::csdE | pNZ44 containing gene csdE | This work |
| pNZ44::csdF | pNZ44 containing gene csdF | This work |
| pNZ44::csdEF | pNZ44 containing genes csdE and csdF | This work |
| Bacteriophages | ||
| jj50* | Spontaneously acquired derivative of jj50 (NC_008371.1), propagated on NZ9000 | This work |
| p2 | 936 species, propagated on NZ9000 | Ref. 48 |
| sk1 | 936 species, propagated on NZ9000 | Ref. 49 |
| MCC1 | Derivative of sk1, propagated on NZ9000 | Ref. 8 |
| LC3 | P335 species, propagated on 3107 | Ref. 50 |
| TP901-1 | P335 species, propagated on 3107 | Ref. 47 |
| 66901 | 936 species, propagated on 3107 | Ref. 51 |
| 66902 | 936 species, propagated on 3107 | UCC Culture Collection |
| 66903 | 936 species, propagated on 3107 | UCC Culture Collection |
| 62604 | 936 species, propagated on 3107 | Ref. 51 |
| 62605 | 936 species, propagated on 3107 | Ref. 51 |
| 62601 | 936 species, propagated on 3107 | Ref. 51 |
| 63301 | P335 species, propagated on 3107 | Ref. 52 |
| 50101 | P335 species, propagated on 3107 | Ref. 52 |
| 07501 | P335 species, propagated on 3107 | Ref. 53 |
| 58601 | P335 species, propagated on 3107 | Ref. 53 |
| 86501 | P335 species, propagated on 3107 | Ref. 52 |
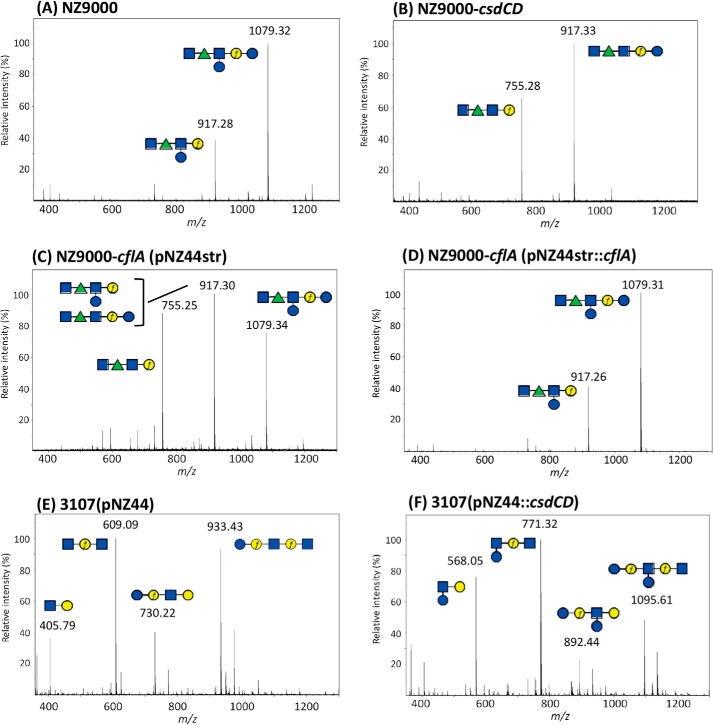
CWPS produced by each of these constructed strains was extracted by HF treatment, and the obtained rhamnan and PSP oligosaccharides were separated by size-exclusion chromatography (SEC-HPLC) (9). The purified components were hydrolyzed, and their monosaccharide composition was determined, whereas they were also analyzed by MALDI-TOF MS. Compositional analysis of the PSP of the L. lactis NZ9000-csdCD double mutant revealed a clear decrease of Glc/Gal ratio compared with that of L. lactis NZ9000 (Table 2). Structural modification of the PSP from L. lactis NZ9000-csdCD was confirmed by MALDI-TOF MS analysis. The obtained mass spectrum of the WT L. lactis NZ9000 purified PSP oligosaccharides exhibits a major peak corresponding to the hexasaccharide subunit in agreement with the structure previously established by NMR (11) (Fig. 3A), whereas this peak was absent in the spectrum of the PSP extracted from the L. lactis NZ9000-csdCD mutant. The latter exhibits a major peak that corresponds to a pentasaccharide subunit resulting from the loss of one hexose (Hex) (Fig. 3B). Methylation analysis showed that terminal Glc (t-Glc) detected in the WT PSP was absent in the PSP of mutant NZ9000-csdCD, thus confirming that the residue absent in the PSP of NZ9000-csdCD was the side-chain Glc. To examine the involvement of the individual csdC and csdD genes, the structure of PSP purified from the single csdC and csdD mutants was also analyzed. Monosaccharide composition analysis revealed a clear decrease of Glc/Gal ratio in both mutants compared with the PSP of L. lactis NZ9000 (Table 2). MALDI-TOF MS analysis and methylation analysis of the PSP obtained from the corresponding csdD mutant revealed the complete loss of the side-chain Glc, similar to the effect observed in the double csdCD mutant (Fig. S1). However, in the case of the csdC mutant, MS and methylation analyses indicated that a portion of the PSP (estimated at 20% based on methylation analysis) still contained the side-chain Glc despite csdC inactivation (Fig. S1). These results suggest that another enzyme with undecaprenyl-phosphate-glucose (Und-P-Glc) synthase activity partially complements the csdC deficiency by producing the sugar-lipid intermediate that is then available for CsdD. Altogether, these results demonstrate that the csdCD gene pair is involved in Glc side-chain addition on the PSP subunit .
Table 2.
Relative monosaccharide composition of PSP oligosaccharides purified by HP-SEC after HF extraction from the different L. lactis strains
| Strain | Rha | GlcNAc | Gala | Glc |
|---|---|---|---|---|
| L. lactis NZ9000 | 0.8 | 2.6 | 1 | 2.4 |
| L. lactis NZ9000-csdCD | 0.6 | 2.1 | 1 | 1 |
| L. lactis NZ9000-csdC | 0.5 | 2.1 | 1 | 1.1 |
| L. lactis NZ9000-csdD | 0.6 | 2.5 | 1 | 1.1 |
| L. lactis NZ9000-cflA | 0.7 | 2 | 1 | 1.3 |
| L. lactis 3107 pNZ44 | 0 | 1.1 | 1 | 0.4 |
| L. lactis 3107 pNZ44::csdCD | 0.3b | 1.0 | 1 | 0.9 |
a Values are standardized relative to Gal.
b Presumed contamination of PSP by short rhamnan chains during HP-SEC purification.
Figure 3.
MALDI-TOF MS spectra obtained on purified PSP oligosaccharides extracted by HF treatment from cell walls of WT L. lactis NZ9000 (A), L. lactis NZ9000-csdCD mutant (B), L. lactis NZ9000-cflA(pNZ44str) flippase mutant (C), L. lactis NZ9000-cflA(pNZ44str::cflA) complemented flippase mutant (D), L. lactis 3107 with empty pNZ44 (E), and L. lactis 3107(pNZ44::csdCD) (F). m/z values on spectra correspond to [M + Na]+ ion adducts. During HF extraction, the polymeric PSP of L. lactis NZ9000 is cleaved at the level of phosphodiester bonds, leading to hexasaccharide (Mcalc 1079.37) and also partially cleaved after Galf, leading to a pentasaccharide (Mcalc 917.32), as shown previously by NMR (9). Similarly, the polymeric PSP of L. lactis 3107 is cleaved during HF extraction at the level of phosphodiester bonds, leading to pentasaccharide (Mcalc 933.32), and also partially after the two Galf present in the structure, leading to fragments with Mcalc of 730.24, 609.21, and 406.13 (for Na+ adducts) (10). Calculated and observed m/z values of the different oligosaccharide structures tentatively assigned to major peaks of the spectra are listed in Table S2. Structures are drawn using the symbol nomenclature for graphical representation of glycans (SNFG). Blue square, GlcNAc; green triangle, Rha; blue circle, Glc; yellow circle, Gal. f, furanose.
In the L. lactis 3107 strain expressing functional csdCD, compositional analysis of the purified PSP revealed an increase of Glc relative to Gal compared with the value in the PSP of WT L. lactis 3107 (Table 2). MALDI-TOF MS analysis confirmed the presence of PSP with an additional hexose in L. lactis 3107(pNZ44::csdCD) compared with WT 3107 (Fig. 3, E and F). The presence of the Glc side chain in L. lactis 3107(pNZ44::csdCD) was confirmed by methylation analysis.
CsdAB is required for rhamnan glucosylation
To assess the function of CsdAB, a similar strategy to the one employed for CsdCD was followed, consisting of mutant/derivative construction in L. lactis NZ9000 and 3107 followed by CWPS analysis. First, in L. lactis NZ9000-csdAB, PSP monosaccharide composition was unchanged compared with WT (data not shown). Because the rhamnan component purified from L. lactis NZ9000 contains mainly Rha and only trace amounts of Glc, Gal, and GlcNAc, the influence of csdAB inactivation on rhamnan composition could not be clearly assessed. Therefore, the csdAB genes were overexpressed in L. lactis NZ9000 to investigate their role.
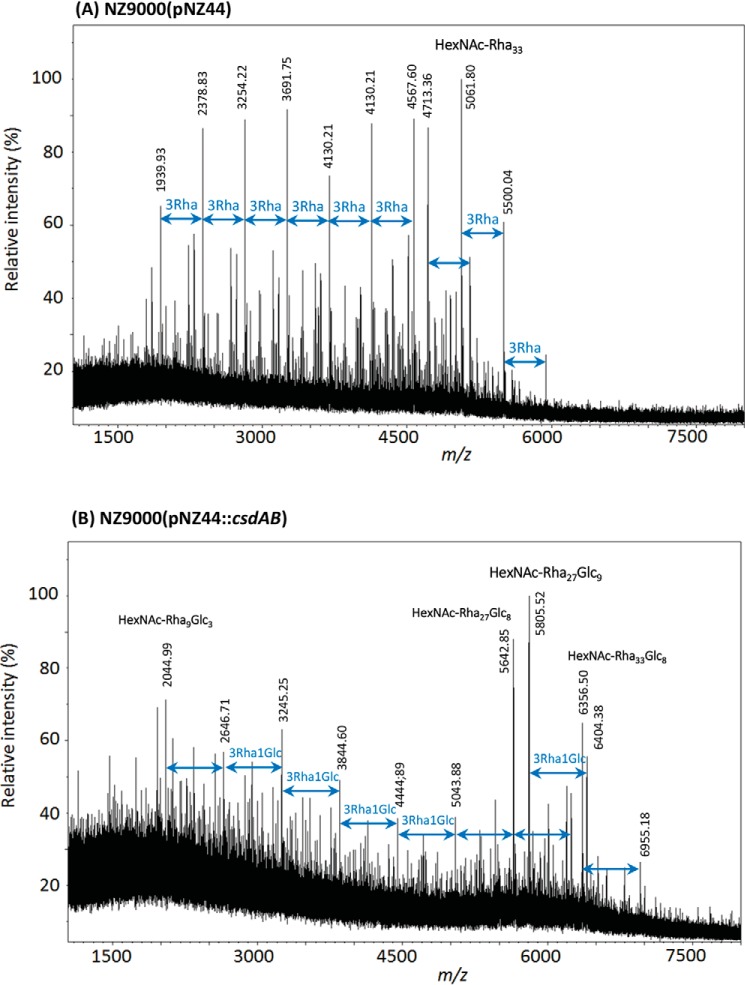
We observed that csdAB overexpression in L. lactis NZ9000 caused a substantial increase of Glc in the rhamnan component (Glc/Rha ratio around 1:3), whereas just a low amount of Glc, relative to Rha, was detected in the WT rhamnan. MALDI-TOF MS analysis of rhamnan showed a complex pattern, with higher m/z value peaks in csdAB-overexpressing NZ9000 (pNZ44::csdAB) compared with NZ9000 (WT) (Fig. 4). In the spectrum of WT rhamnan, a major family of signals were separated by 438 mass units (Fig. 4A), which can be attributed to a difference of one rhamnan subunit constituted of 3 Rha (9). In contrast, in the spectrum of the rhamnan from csdAB-overexpressing NZ9000, a major family of signals was detected with 600-mass unit increments that can be assigned to a difference of 3 Rha plus 1 Hex, corresponding to one rhamnan subunit bearing a Glc side chain (Fig. 4B). Methylation analysis confirmed a substantial increase of t-Glc in the rhamnan purified from L. lactis NZ9000 (pNZ44::csdAB), whereas a 2,3-disubstituted Rha appeared with a concomitant decrease in the level of 3-Rha, compared with the control NZ9000 rhamnan (Fig. S2). These results confirmed the presence of Glc substitutions on rhamnan chains and identified the site of branching. From the methylation analysis, we deduced the following structure for the subunits substituted with Glc: →2)-α-Rha-(1→2)-α-Rha-(1→3)-[Glc-(1→2)]-α-Rha-. No effect of csdAB overexpression was detected on PSP structure either by composition or MS analysis (Fig. S3).
Figure 4.
MALDI-TOF MS spectra of rhamnan purified from L. lactis NZ9000(pNZ44) (A) and L. lactis NZ9000(pNZ44::csdAB) (B). All m/z values correspond to [M + Na]+ adducts.
Overexpression of csdA alone in NZ9000 did not modify the rhamnan or PSP structures (Figs. S3B and S4B). In contrast, overexpression of csdB alone in NZ9000 was shown to result in an increase of Glc in the rhamnan component (Glc/Rha ratio around 1:4) and an MS spectrum showing higher molecular mass species (Fig. S4C) similar to that observed for csdAB overexpression. Interestingly, the MS spectrum of PSP from NZ9000 overexpressing only csdB also appeared modified, with an increased intensity of the peak corresponding to the pentasaccharide versus the hexasaccharide peak (Fig. S3C). This result was corroborated by methylation analysis indicating a reduction of Glc side-chain addition in PSP in this latter strain. These results can be explained by assuming that part of the Und-P-Glc intermediate synthesized by CsdC, which possesses the same Und-P-Glc synthase activity as CsdA and is involved in PSP glucosylation, is titrated away by CsdB when this latter protein is overexpressed and is directed to perform rhamnan glucosylation.
csdAB genes from NZ9000 were then introduced on plasmid pNZ44::csdAB in strain 3107, which lacks functional homologs, to evaluate their role and significance for this strain. The composition of the purified rhamnan component of the CWPS was shown to exhibit a very substantial increase of Glc/Rha ratio (around 1:2), whereas only traces of Glc were found in the rhamnan peak of control L. lactis 3107 pNZ44. MALDI-TOF MS analysis confirmed a clear mass increase for the rhamnan extracted from L. lactis 3107 pNZ44::csdAB compared with the control L. lactis 3107 pNZ44 and WT strains (Fig. S5). When either csdA or csdB was expressed individually in L. lactis 3107, no difference in rhamnan composition or MS spectrum was observed (data not shown), indicating that csdA and csdB are both required for rhamnan glucosylation in strain 3107. Together, these results imply that CsdA and CsdB are functional when heterologously expressed in L. lactis 3107.
CsdEF is involved in LTA galactosylation
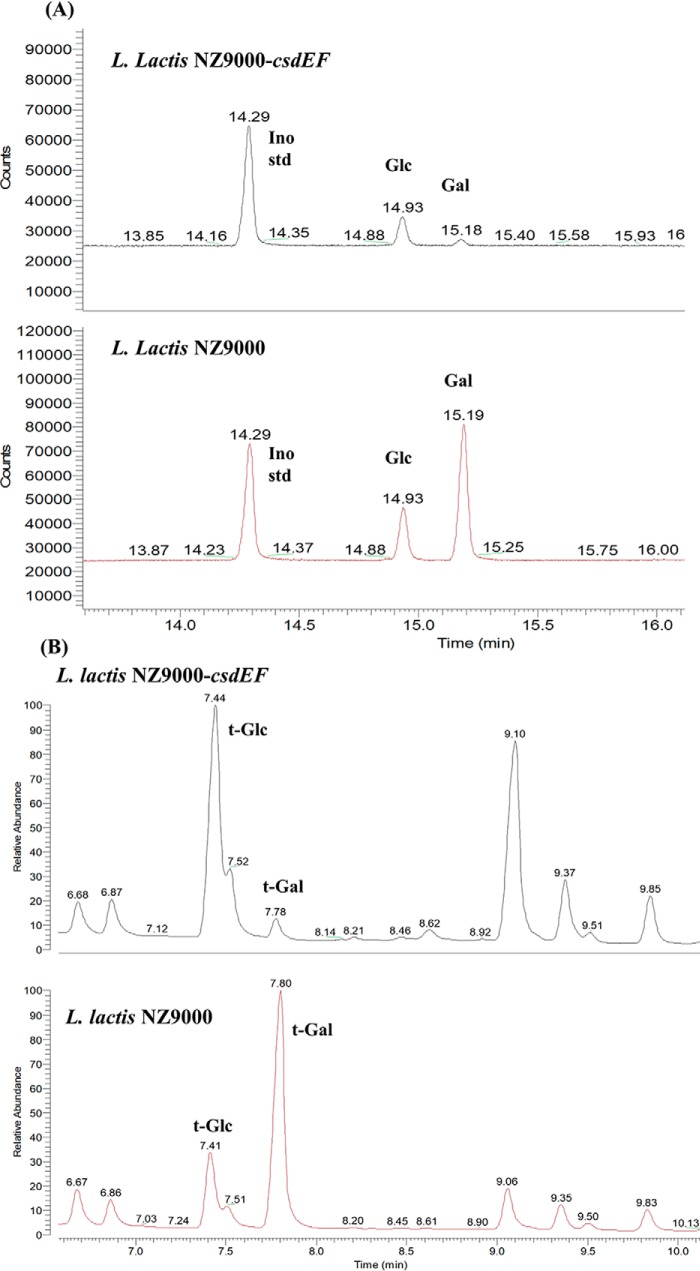
No modification of the rhamnan or PSP was detected in L. lactis NZ9000-csdEF or in L. lactis NZ9000 pNZ44::csdEF (data not shown). These results suggest that CsdE and CsdF are involved in the glycosylation of an alternative cell wall glycopolymer, such as WTA or LTA. Of note, L. lactis has previously been shown to synthesize LTA, which consists of glycerol-phosphate chains, with Gal as a sugar substituent (15). To assess whether LTA glycosylation depends on the products of csdEF, LTA was extracted from L. lactis NZ9000 and its mutant NZ9000-csdEF with hot phenol (16). The degree of LTA galactosylation was assessed based on relative quantities of glycerol, Gal, and terminal Gal (t-Gal) in two preparations. Our results demonstrated a 10-fold decrease in Gal levels at constant glycerol levels in the L. lactis NZ9000-csdEF mutant preparation (Fig. 5A). In agreement with these results, the amount of t-Gal decreased dramatically in this sample, compared with the preparation from L. lactis NZ9000 (Fig. 5B). These data clearly indicate that mutating both csdE and csdF negatively affects galactosylation of poly(glycerophosphate) chains of LTA in L. lactis NZ9000.
Figure 5.
A, monosaccharide analysis of crude phenol extracts from L. lactis NZ9000 and NZ9000-csdEF. Each extract (5 mg), along with Ino (90 μg) as an internal standard, were treated with HF, hydrolyzed, converted into alditol acetates, and analyzed by GC with flame-ionization detection. B, methylation analysis of crude phenol extracts from L. lactis NZ9000 and NZ9000-csdEF. Samples were analyzed by GC-MS. The figures were zoomed onto the region corresponding to hexoses.
The cflA gene encoding a putative flippase is involved in glucosylation of CWPS components as well as LTA galactosylation
As previously proposed (5), a protein with flippase activity should be involved in the TGS pathway. A gene (cflA) encoding a putative flippase with sequence identity to the L. monocytogenes GtcA flippase (7) was identified in the L. lactis NZ9000 genome. Analysis of the PSP oligosaccharides purified from an L. lactis NZ9000-cflA mutant revealed a decreased level of Glc in PSP compared with the WT, suggesting incomplete addition of Glc side chain onto PSP subunits (Table 2). MS analysis confirmed this observation with a modified spectrum of the mutant strain versus the WT, in agreement with the presence of PSP oligosaccharides devoid of side-chain Glc (Fig. 3C). Complementation of the cflA mutant allowed the phenotypic restoration of the WT PSP structure as shown by MALDI-TOF MS analysis of PSP oligosaccharides (Fig. 3D).
To investigate the role of cflA in Glc substituent addition onto rhamnan, the csdAB genes were overexpressed (employing plasmid pNZ44str::csdAB) in the L. lactis NZ9000-cflA mutant. Whereas, as described above, overexpression of csdAB in L. lactis NZ9000 led to a very high level of Glc grafted to rhamnan as observed by composition analysis (Glc/Rha ratio around 1:4), NZ9000-cflA pNZ44str::csdAB showed no (or very little) Glc substitution of the rhamnan. This effect could also be revealed by MALDI-TOF MS analysis. Whereas in the NZ9000 background, overexpression of csdAB lead to a significant increase of the rhamnan mass as described above, in the cflA background, overexpression of csdAB had no effect (Fig. S4, E and F). These results clearly show that CflA is required for glucosylation of rhamnan. Our results therefore identify CsdA, CsdB, and CflA as a three-component rhamnan glucosylation system in L. lactis.
The effect of cflA inactivation was also evaluated on LTA galactosylation. Monosaccharide analysis of dephosphorylated phenol extracts of the L. lactis NZ9000-cflA mutant, compared with the WT strain showed the absence of any detectable Gal. Methylation analysis of this preparation did not reveal any terminal Gal, confirming that the LTA of this strain lacked Gal substitution. Complementation of the cflA mutant restores the level of Gal and t-Gal (Fig. S6). These results indicate that the flippase CflA acts in concert with CsdE and CsdF in the galactosylation process of LTA chains.
Derivative strain fitness and bacteriophage interactions
Phenotypic effects due to the mutation or overexpression of csdAB, csdCD, and csdEF were investigated. All derived L. lactis NZ9000 strains were grown under optimal as well as various stress conditions by incorporating growth-limiting concentrations of NaCl, ethanol, and EDTA and reduced pH. It was observed that under optimal growth conditions (in M17Glc broth), nonsense mutations and overexpression of these three sets of genes did not exert any detrimental effect on growth rate (data not shown). With the exception of the antimicrobial peptide nisin, the stressors appeared to similarly affect both the WT and the derived strains. Overexpression of the csdEF provided increased resistance to nisin, whereas the opposite effect was observed for the nonsense mutant (Fig. 6). Interestingly, when the effect of a nonsense mutation in cflA was investigated, it was observed that the mutant strain exhibited a reduced growth rate compared with the control strain even under optimal conditions (Fig. S7).
Figure 6.
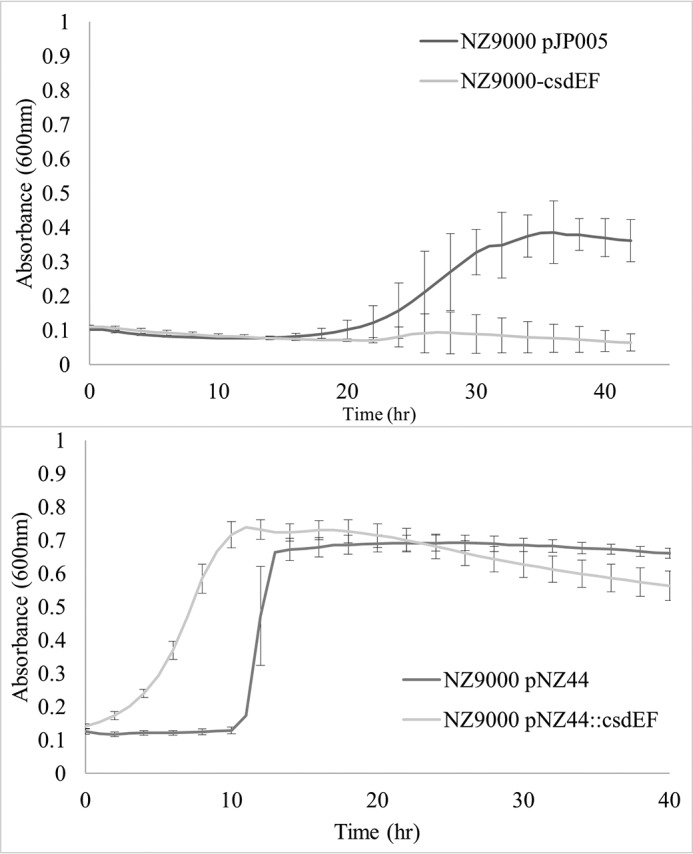
Growth behavior of L. lactis NZ9000 controls and csdEF derivatives in M17 Glc and 15 μg/ml nisin. Error bars, S.D.
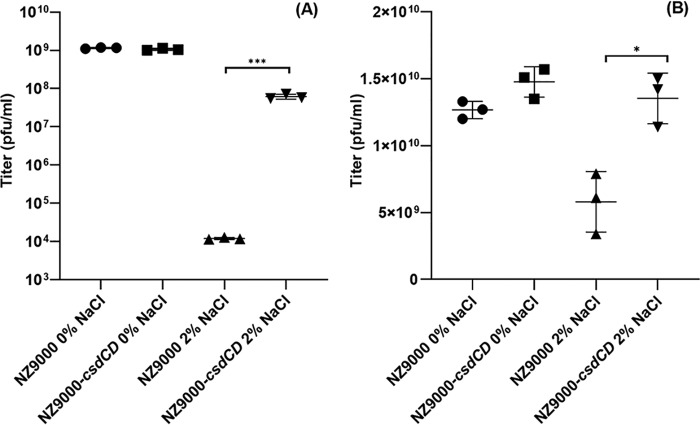
Lactococcal PSP is required for adsorption by certain phages, and mutations that impair or alter PSP biosynthesis may lead to altered phage sensitivities (12–14). These findings combined with our observation that CsdCD is responsible for PSP glucosylation prompted us to determine whether this Glc substitution alters phage-host interactions. The mutant strain L. lactis NZ9000-csdCD was assayed for its sensitivity to the phages (936-type phages) listed in Table 1. The presence of the double knockout mutation did not significantly affect the infective capabilities of the phages under standard plaquing conditions (data not shown). Several environmental conditions that more accurately reflect the suboptimal environments in which phage-host interactions may be taking place, particularly those conditions associated with dairy fermentations, were also tested (see “Experimental procedures”). Among these conditions, significantly altered phage sensitivities between L. lactis NZ9000 and L. lactis NZ9000-csdCD were observed under high-salt conditions. Fig. 7 indicates the much higher efficiency (3.7 log) by which phage p2 infects the derivative strain L. lactis NZ9000-csdCD when compared with p2 infection of the L. lactis NZ9000 in the presence of salt, a difference that is not observed under standard conditions. A similar but much more subtle effect is observed for phage sk1. In addition, phage adsorption assays revealed that p2 exhibits enhanced reversible and irreversible adsorption to L. lactis NZ9000-csdCD relative to L. lactis NZ9000 under standard laboratory conditions (Fig. S8).
Figure 7.
Titer of phages p2 (A) and sk1 (B) against L. lactis NZ9000 and L. lactis NZ9000-csdCD in 0 and 2% NaCl environment. Each data point represents a biological replicate. p values are indicated by asterisks. ***, p < 0.0001; *, p < 0.05. Error bars, S.D.
The structural modifications incurred by the changes in expression of csdAB and csdEF and their effect on phage/host interactions were also examined. L. lactis NZ9000-csdAB, NZ9000-csdEF, NZ9000 pNZ44::csdAB, NZ9000 pNZ44::csdEF, and L. lactis 3107 pNZ44::csdEF did not exhibit altered phage sensitivity profiles (four 936-type phages with L. lactis NZ9000 as a host and six 936-type and seven P335-type phages with L. lactis 3107 as a host; Table 1). In contrast, overexpression of csdAB in L. lactis 3107 caused reduced sensitivities against phages that belong to either the P335 or 936 group (Fig. S9A). Similar results were observed when csdAB was overexpressed in L. lactis VES5751, a PSP-deficient derivative of L. lactis MG1363 that is resistant to infection by most phages commonly infecting the WT strain (11). Two phages, jj50* and MCC1, derivatives of phage jj50 (NC_00837) and sk1 (NC_001835.1), respectively, are unaffected by the PSP-deficient phenotype of L. lactis VES5751 and can maintain their ability to infect it. However, upon overexpression of csdAB in L. lactis VES5751, the infectivity of jj50* is reduced approximately 100-fold compared with the control strains, whereas MCC1 appears to be unaffected by csdAB overexpression (Fig. S9B).
Finally, the effect of mutation of the flippase gene, cflA, on bacteriophage interaction was investigated. Surprisingly, despite the effect that the mutation has on the growth of the strain (Fig. S7), the tested phages were equally efficient at infecting the control and mutant L. lactis NZ9000-cflA strains, possibly owing to the maintenance of the Glc side chain in the PSP of L. lactis NZ9000-cflA, albeit at a reduced frequency compared with the WT strain.
Discussion
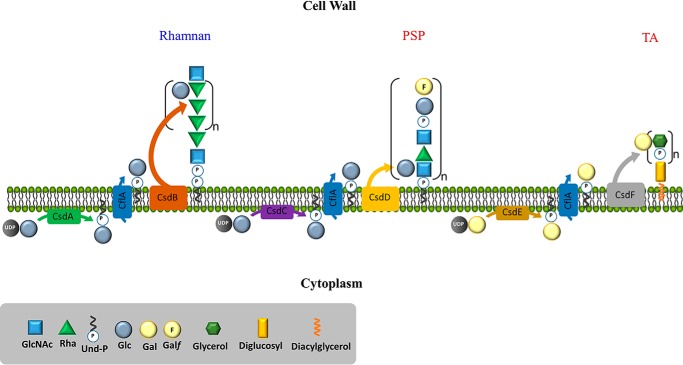
In the current work, we identified three gene pairs in L. lactis located outside the cwps gene cluster, each encoding two predicted glycosyltransferases, one GT able to transfer sugar from NDP-sugar to Und-P and the second a PolM GT, which possesses the characteristics of GTs involved in extracytoplasmic modification of bacterial glycoconjugates. We successfully assigned a function for these three gene pairs, showing that each is involved in the glycosylation of a distinct glycopolymer of the lactococcal cell wall. The csdEF gene pair is the most commonly encountered set of the three, whereas the complete absence of any intact csd gene (as seen in L. lactis JM1/JM2) hints at the accessory function of the gene pair products. In addition, our results clearly indicate that the three glycosylation systems share a common flippase involved in transferring Und-P-Glc as well as Und-P-Gal from the inner side to the outer side of the cytoplasmic membrane.
Original work involving the pathogenic microorganism Shigella flexneri first described the mechanism by which the O-antigen of the lipopolysaccharide is glycosylated and how this modified the strain's virulence and resistance to innate immune killing. This glycosylation process was shown to be mediated by the function of three contiguously located genes, gtrABV (17, 18). Similar TGSs (5) have been implicated in the serovar conversion of pathogenic species such as Listeria monocytogenes (19) as well as many Salmonellae (20). Our results show that the csdCD gene pair in L. lactis forms part of such a TGS and is involved in the attachment of the Glc side chain on PSP subunits. In addition, these genes are also functional when expressed in L. lactis 3107 (which lacks a functional homolog of csdC). We propose that CsdC is involved in the transfer of Glc from UDP-Glc to Und-P at the inner face of the cytoplasmic membrane, whereas CsdD is presumed to catalyze Glc transfer onto PSP outside the cytoplasmic membrane (Fig. 8). Interestingly, CsdD is absolutely required for PSP glucosylation. In contrast, in the absence of CsdC, partial glucosylation of PSP was still observed, indicating that Und-P-Glc substrate can be provided to CsdD by another GT possessing the same activity as CsdC. CsdA described above was also shown to possess Und-P-Glc synthase activity and is thus a likely candidate to fulfill this role. Thus, our results indicate a certain level of functional redundancy and cross-talk among different systems dedicated to glycosylation of different cell wall glycopolymers, mediated by the availability of the Und-P sugar intermediate. We also show that the products of csdAB are responsible for the glucosylation of the rhamnan subunit and that the enzymatic activities of both proteins are required for this addition (Fig. 8). According to methylation analysis, Glc is connected to the 3-Rha of the [-2-α-Rha-2-α-Rha-3-α-Rha-] subunit. Overexpression of csdB, which encodes a PolM GT in NZ9000, allowed grafting of one Glc side chain on almost every 3-Rha subunit, as was observed upon overexpression of csdAB. These results suggest that CsdB can utilize the Und-P-Glc substrate produced by another Und-P-Glc synthase apart from that produced by its cognate partner CsdA, similar to our observations for the CsdCD pair. These observations confirm that CsdB provides specificity of the glucosylation reaction toward rhamnan. Finally, the gene pair csdEF was shown to be required for LTA galactosylation (Fig. 8). TGSs have previously been shown to glycosylate polymers derived from both a Wzy-dependent and ATP-binding cassette (ABC) transporter assembly pathway (5, 21). However, the existence of three TGSs to modify three distinct glycopolymers within a single bacterial species has not previously been observed.
Figure 8.
Schematic representation of the suggested functions of the proteins encoded by each of the three identified gene pairs (csdAB, csdCD, and csdEF) and the flippase-encoding gene cflA.
With respect to gene organization, a hallmark of many TGSs is the close genomic proximity of the genes involved in glycosylation (18, 21). Interestingly, a gene encoding a putative flippase was identified on the L. lactis NZ9000 genome (cflA) that is in a noncontiguous position with respect to the gene pairs highlighted above. Inactivation of cflA was shown to completely block glucosylation of the rhamnan as well as galactosylation of LTA and, although partially, to block PSP glucosylation. From these results, it is clear that CflA is shared by the three TGSs with CsdAB, CsdCD, and CsdEF (Fig. 8). Partial substitution of PSP subunits was still present in the cflA mutant, suggesting the presence of another flippase endowed with the same activity as CflA, although a sequence homology search did not return any likely candidate. Interestingly, unlike the mutants of csdAB or csdCD, the cflA mutant strain exhibits a distinctly reduced growth rate, which may result from an indirect effect of the accumulation of Und-P-Glc intermediates preventing recycling of Und-P (required for essential processes such as peptidoglycan synthesis).
It has previously been established that the receptor for both 936 (10, 13, 22–24) and P335 group phages (10, 12, 25) is saccharidic in nature. It was thus expected that modifications of the lactococcal CWPS affect phage infectivity. We observed that phage p2 preferentially infected the csdCD mutant derivative under saline conditions. Such environmental conditions seemingly destabilize the already weak protein-saccharide interactions characteristic between the 936-type receptor-binding proteins (RBPs) and their CWPS receptors. Because phage p2 infects and also adsorbs more efficiently to the derivative lacking the PSP glucose side chain, we hypothesize that its RBP has evolved to preferentially bind to the primary pentasaccharide unit of the PSP. The existence of the Glc side chain on PSP may thus provide a degree of phage insensitivity to the strain. Similar effects from the decoration of cell wall polysaccharide components, such as WTA, have previously been reported for phage adsorption on various Gram-positive bacterial strains (3). The overexpression of csdAB and subsequent increase of Glc side chain on rhamnan was shown to also act as a phage predation deterrent both in L. lactis 3107 (PSP(+) and rhamnan-unexposed) and L. lactis VES5751 (PSP(−) and rhamnan-exposed).
In addition to phage/host interactions, cell wall glycopolymers and their substitutions may contribute to increased antibiotic resistance (15, 26, 27). Here, we investigated the effect of LTA glycosylation on various phenotypic responses in L. lactis. Interestingly, our results support previous evidence for the mechanism of nisin resistance in L. lactis through increased TA substitution and increased septal density (15). In detail, altering the levels of galactosylation of the teichoic acids in L. lactis NZ9000 led to equivalent changes in nisin resistance (i.e. increased glycosylation caused increased nisin resistance and vice versa).
Three-component glycosylation systems are commonly associated with prophage elements in Shigella (18) and Salmonella species (28). They have also been shown to directly interfere with phage/host interactions in L. monocytogenes (7, 19). Interestingly, we observed that the genomic vicinity of csdCD is rich in mobile elements, including transposases and prophage components. This observation combined with the effect on phage-host interactions upon modulation of the gene pair expression strongly suggests that such mobile gene cassettes provide a swift response to phage predation by adding nonessential decorations onto an already existing polysaccharide structure. We speculate that genes encoding such CWPS-modulating functions have been “hijacked” by active prophages to further enhance any pre-existing superinfection exclusion mechanisms and activities. Our research reiterates the structural complexity as well the dynamic nature of the CWPS in L. lactis. Such characteristics were previously thought to stem from the genomic variability of the central cwps gene cluster yet are now also seen to be influenced by additional genetic components beyond the cwps cluster. However, further analysis and investigation should be performed to evaluate the phenotypic implications of such modifications of cell wall glycopolymers.
Experimental procedures
Bacterial strains, phages, and growth conditions
Bacterial strains, plasmids, and phages used in this study are listed in Table 1. Strains were grown at 30 °C overnight in M17 broth and/or M17 agar (Oxoid Ltd.) supplemented with 5 g/liter glucose. Chloramphenicol (5 μg/ml), erythromycin (5 μg/ml), and tetracycline (10 μg/ml) (Sigma–Aldrich) were added to the media where appropriate. Induction of genes placed under the nisin-controlled promoter, Pnis, was achieved through supplementation of the media with nisaplin (40 ng/ml) (DuPont).
Bacteriophage assays
Phages used in this study are listed in Table 1. Propagation of phages on their respective host strains was performed as described previously (29). Similarly, both spot/plaque assays (30) and adsorption assays (31) were performed as described previously with sodium chloride added to a final concentration of 1–2% (w/v), where indicated.
Bioinformatic analysis
Candidate genes encoding proteins responsible for glucose side-chain decoration of the PSP of L. lactis MG1363 and L. lactis SMQ-388 were selected based on amino acid homologies to proteins previously investigated in L. monocytogenes (7). To this end, the proteins indicated in Fig. 1 were compared using BLASTP against the compiled coding sequences (CDS) of the L. lactis MG1363 genome (GenBankTM accession number NC_009004.1). Genes in the L. lactis MG1363 genome, which are not associated with the cwps gene cluster, were selected for further investigation based on their corresponding amino acid similarity to the following L. monocytogenes proteins: GtcA; GtcB and GtlA; and GtcC and GtlB (6, 7). Selected gene sequences from the L. lactis MG1363 genome were used to search for homologues in the genomes of L. lactis 3107 and L. lactis SMQ-3886 using BLASTN. Intrinsic properties of protein sequences were assessed using HHPred (32) and TMHMM online predictive tools (33, 34).
Selected genes were analyzed based on sequence identity against all publicly available L. lactis chromosomal and plasmid sequences, whereas protein sequence comparisons were performed using all-against-all bidirectional BLAST alignments (35) (cut-off: E-value 0.0001, with at least 50% identity across at least 50% of either protein sequence). A heat-map matrix, with the presence/absence/nonfunctionality of each of the retrieved candidate genes from all assessed L. lactis strains, was created and run in MeV suite (version 4.9) (36).
Molecular cloning
All recombinant plasmids (Table 1) were generated in L. lactis NZ9000, and primers used can be seen in Table S1 (Eurofins MWG). The gene pairs of interest, csdAB, csdCD, and csdEF, were each amplified and cloned in the high-copy number constitutive expression vector pNZ44 (37). Due to the presence of pJP005 in L. lactis NZ9000-cflA that carries a chloramphenicol resistance marker, csdAB was also separately cloned into pNZ44str, a pNZ44-derivative that carries a streptomycin resistance marker instead of the chloramphenicol resistance marker, using the same oligonucleotides. Likewise, each of the genes from the two pairs, csdAB and csdEF, was cloned independently into the pNZ44 vector.
Recombineering and CRISPR-assisted recombineering
Recombineering was performed as described previously (38–40) with modifications to the assay to allow use with L. lactis NZ9000 on the selected genes. CRISPR-Cas9–assisted recombineering was adapted from a previous publication (41) and was used to create a knockout mutant of the cflA gene because the above-mentioned recombineering protocol proved to be unsuccessful, possibly due to decreased fitness of such mutants. A novel plasmid, named pCNR, containing the replication genes repA, repD, and repE from the backbone of the plasmid pPTPi (42) along with the chloramphenicol resistance gene (cmr), PnisA promoter, and the ssDNA-binding protein–encoding gene, recT, from plasmid pJP005 (39), was constructed. Subsequently, the single-step approach to CRISPR-Cas9–assisted recombineering was employed as outlined previously (42).
CWPS and LTA extraction, purification, and analysis
CWPS was extracted from cell envelope fractions prepared from L. lactis cells and analyzed as described previously (9). Briefly, cell walls were prepared and treated with HF to extract CWPS. Rhamnan and PSP oligosaccharides present in the HF extract were further separated by SEC-HPLC with two columns in tandem (Shodex Sugar KS-804 and KS-803 columns, Showa Denko, Japan) with a refractometer (2414 Refractive Index Detector, Waters) and/or UV detector at 206 nm. Fractions corresponding to peaks containing rhamnan and PSP oligosaccharides were collected and dried under vacuum. Monosaccharide composition was determined by high-performance anion-exchange chromatography coupled with pulse-amperometric detection (ICS5000 system, Thermo Fisher Scientific). Purified fractions were analyzed by MALDI-TOF MS using 2,5-dihydroxybenzoic acid matrix with an UltrafleXtreme instrument (Bruker Daltonics, Bremen, Germany). MS data are accessible on the Glycopost website (54), ID: GPST000056.
Crude LTA was extracted essentially as described previously (16). Identical amounts (2–5 mg) of LTA extracts of the WT strain L. lactis NZ9000 and mutant L. lactis NZ9000-csdEF were treated with HF (48%, 4 °C, 24 h) and subjected to comparative analysis for glycerol and monosaccharides, as well as methylation analysis. m-Inositol (Ino) was used as an internal standard to assess glycerol content. The use of phenol for the isolation of LTAs should guarantee relative purity of the sample, although the possibility of cross-contamination from other TA structures, such as WTA, cannot be excluded.
Methylation analysis was performed using the Ciucanu–Kerek procedure (43) and modified by Read et al. (44). The product was hydrolyzed with 4 m TFA (110 °C, 3 h), dried, reduced with NaBD4, converted into the alditol acetates by conventional methods, and analyzed by GC-MS (45).
Strain fitness, viability test, and biofilm formation
Viability/survival and fitness of strains derived from L. lactis NZ9000 were investigated following growth under various stress-inducing conditions. Initially, the level of halotolerance (5.5% (w/v) NaCl), ethanol tolerance (8% (v/v) EtOH), tolerance to low pH conditions (pH 4.8), chelating agents (750 μm EDTA), and the bacteriocin nisin (15 μg/ml) was determined for L. lactis NZ9000. Derived mutant strains (Table 1) were also similarly tested. L. lactis NZ9000-cflA (Table 1) was evaluated to compare its growth behavior with that of the control strain L. lactis pVPL3004/pCRISPR.
Data availability
All data created, with the exception of MS data, can be found within the document. MS data have been deposited and are available on the Glycopost depository (54) with the ID GPST000056.
Author contributions
I. T., P. C., J. M., M.-P. C.-C., and D. v. S. conceptualization; I. T., P. C., I. S., S. P., F. F., and M.-P. C.-C. data curation; I. T., P. C., I. S., S. P., F. F., and M.-P. C.-C. formal analysis; I. T., P. C., and M.-P. C.-C. investigation; I. T., P. C., I. S., and M.-P. C.-C. methodology; I. T. writing-original draft; I. T., P. C., I. S., S. P., F. F., J. M., M.-P. C.-C., and D. v. S. writing-review and editing; J. M., M.-P. C.-C., and D. v. S. supervision; J. M. and D. v. S. funding acquisition; M.-P. C.-C. and D. v. S. project administration.
Supplementary Material
Acknowledgment
Prof. S. Moineau is acknowledged for kindly providing the genomic sequence for the strain L. lactis SMQ-388.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S9.
S. Moineau, personal communication.
- LTA
- lipoteichoic acid
- CWPS
- cell wall polysaccharide(s)
- GT
- glycosyltransferase
- PolM GT
- polytopic glycosyltransferase
- PSP
- polysaccharide pellicle
- RBP
- receptor-binding protein
- SEC
- size-exclusion chromatography
- TGS
- three-component glycosylation system
- TMH
- transmembrane helix
- Und-P
- undecaprenyl-phosphate
- WTA
- wall teichoic acid
- HF
- hydrogen fluoride
- TA
- teichoic acid
- Gal
- galactose
- Glc
- glucose
- t-Glc
- terminal Glc
- t-Gal
- terminal Gal
- Hex
- hexose
- Rha
- rhamnose
- ABC
- ATP-binding cassette
- Ino
- m-inositol.
References
- 1. Chapot-Chartier M. P., and Kulakauskas S. (2014) Cell wall structure and function in lactic acid bacteria. Microb. Cell Fact. 13, S9 10.1186/1475-2859-13-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Percy M. G., and Gründling A. (2014) Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu. Rev. Microbiol. 68, 81–100 10.1146/annurev-micro-091213-112949 [DOI] [PubMed] [Google Scholar]
- 3. Brown S., Santa Maria J. P. Jr., and Walker S. (2013) Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mistou M. Y., Sutcliffe I. C., and van Sorge N. M. (2016) Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in gram-positive bacteria. FEMS Microbiol. Rev. 40, 464–479 10.1093/femsre/fuw006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mann E., and Whitfield C. (2016) A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can. J. Chem. 94, 883–893 10.1139/cjc-2015-0594 [DOI] [Google Scholar]
- 6. Rismondo J., Percy M. G., and Gründling A. (2018) Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J. Biol. Chem. 293, 3293–3306 10.1074/jbc.RA117.001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spears P. A., Havell E. A., Hamrick T. S., Goforth J. B., Levine A. L., Abraham S. T., Heiss C., Azadi P., and Orndorff P. E. (2016) Listeria monocytogenes wall teichoic acid decoration in virulence and cell-to-cell spread. Mol. Microbiol. 101, 714–730 10.1111/mmi.13353 [DOI] [PubMed] [Google Scholar]
- 8. Theodorou I., Courtin P., Palussière S., Kulakauskas S., Bidnenko E., Péchoux C., Fenaille F., Penno C., Mahony J., van Sinderen D., and Chapot-Chartier M. P. (2019) A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in Lactococcus lactis. J. Biol. Chem. 294, 17612–17625 10.1074/jbc.RA119.009957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadovskaya I., Vinogradov E., Courtin P., Armalyte J., Meyrand M., Giaouris E., Palussière S., Furlan S., Péchoux C., Ainsworth S., Mahony J., van Sinderen D., Kulakauskas S., Guérardel Y., and Chapot-Chartier M.-P. (2017) Another brick in the wall: a rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. mBio 8, e01303–17 10.1128/mBio.01303-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahony J., Cambillau C., and van Sinderen D. (2017) Host recognition by lactic acid bacterial phages. FEMS Microbiol. Rev. 41, S16–S26 10.1093/femsre/fux019 [DOI] [PubMed] [Google Scholar]
- 11. Chapot-Chartier M. P., Vinogradov E., Sadovskaya I., Andre G., Mistou M. Y., Trieu-Cuot P., Furlan S., Bidnenko E., Courtin P., Péchoux C., Hols P., Dufrêne Y. F., and Kulakauskas S. (2010) Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285, 10464–10471 10.1074/jbc.M109.082958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ainsworth S., Sadovskaya I., Vinogradov E., Courtin P., Guerardel Y., Mahony J., Grard T., Cambillau C., Chapot-Chartier M.-P., and van Sinderen D. (2014) Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5, e00880–14 10.1128/mBio.00880-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupont K., Janzen T., Vogensen F. K., Josephsen J., and Stuer-Lauridsen B. (2004) Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 70, 5825–5832 10.1128/AEM.70.10.5825-5832.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farenc C., Spinelli S., Vinogradov E., Tremblay D., Blangy S., Sadovskaya I., Moineau S., and Cambillau C. (2014) Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J. Virol. 88, 7005–7015 10.1128/JVI.00739-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kramer N. E., Hasper H. E., van den Bogaard P. T. C., Morath S., de Kruijff B., Hartung T., Smid E. J., Breukink E., Kok J., and Kuipers O. P. (2008) Increased d-alanylation of lipoteichoic acid and a thickened septum are main determinants in the nisin resistance mechanism of Lactococcus lactis. Microbiology 154, 1755–1762 10.1099/mic.0.2007/015412-0 [DOI] [PubMed] [Google Scholar]
- 16. Sijtsma L., Wouters J. T., and Hellingwerf K. J. (1990) Isolation and characterization of lipoteichoic acid, a cell envelope component involved in preventing phage adsorption, from Lactococcus lactis subsp. cremoris SK110. J. Bacteriol. 172, 7126–7130 10.1128/JB.172.12.7126-7130.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan S., Bastin D. A., and Verma N. K. (1999) Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145, 1263–1273 10.1099/13500872-145-5-1263 [DOI] [PubMed] [Google Scholar]
- 18. West N. P., Sansonetti P., Mounier J., Exley R. M., Parsot C., Guadagnini S., Prévost M.-C., Prochnicka-Chalufour A., Delepierre M., Tanguy M., and Tang C. M. (2005) Optimization of virulence functions through glucosylation of Shigella LPS. Science 307, 1313–1317 10.1126/science.1108472 [DOI] [PubMed] [Google Scholar]
- 19. Eugster M. R., Morax L. S., Hüls V. J., Huwiler S. G., Leclercq A., Lecuit M., and Loessner M. J. (2015) Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 97, 33–46 10.1111/mmi.13009 [DOI] [PubMed] [Google Scholar]
- 20. Davies M. R., Broadbent S. E., Harris S. R., Thomson N. R., and van der Woude M. W. (2013) Horizontally acquired glycosyltransferase operons drive salmonellae lipopolysaccharide diversity. PLoS Genet. 9, e1003568 10.1371/journal.pgen.1003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mann E., Ovchinnikova O. G., King J. D., and Whitfield C. (2015) Bacteriophage-mediated glucosylation can modify lipopolysaccharide O-antigens synthesized by an ATP-binding cassette (ABC) transporter-dependent assembly mechanism. J. Biol. Chem. 290, 25561–25570 10.1074/jbc.M115.660803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tremblay D. M., Tegoni M., Spinelli S., Campanacci V., Blangy S., Huyghe C., Desmyter A., Labrie S., Moineau S., and Cambillau C. (2006) Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188, 2400–2410 10.1128/JB.188.7.2400-2410.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geller B. L., Ngo H. T., Mooney D. T., Su P., and Dunn N. (2005) Lactococcal 936-species phage attachment to surface of Lactococcus lactis. J. Dairy Sci. 88, 900–907 10.3168/jds.S0022-0302(05)72756-9 [DOI] [PubMed] [Google Scholar]
- 24. Mahony J., Kot W., Murphy J., Ainsworth S., Neve H., Hansen L. H., Heller K. J., Sørensen S. J., Hammer K., Cambillau C., Vogensen F. K., and van Sinderen D. (2013) Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl. Environ. Microbiol. 79, 4385–4392 10.1128/AEM.00653-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spinelli S., Campanacci V., Blangy S., Moineau S., Tegoni M., and Cambillau C. (2006) Modular structure of the receptor binding proteins of Lactococcus lactis phages: the RBP structure of the temperate phage TP901-1. J. Biol. Chem. 281, 14256–14262 10.1074/jbc.M600666200 [DOI] [PubMed] [Google Scholar]
- 26. Saar-Dover R., Bitler A., Nezer R., Shmuel-Galia L., Firon A., Shimoni E., Trieu-Cuot P., and Shai Y. (2012) d-Alanylation of lipoteichoic acids confers resistance to cationic peptides in Group B Streptococcus by increasing the cell wall density. PLOS Pathog. 8, e1002891 10.1371/journal.ppat.1002891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giaouris E., Briandet R., Meyrand M., Courtin P., and Chapot-Chartier M.-P. (2008) Variations in the degree of d-alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl. Environ. Microbiol. 74, 4764–4767 10.1128/AEM.00078-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim M., and Ryu S. (2012) Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 86, 411–425 10.1111/j.1365-2958.2012.08202.x [DOI] [PubMed] [Google Scholar]
- 29. Mahony J., McGrath S., Fitzgerald G. F., and van Sinderen D. (2008) Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl. Environ. Microbiol. 74, 6206–6215 10.1128/AEM.01053-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lillehaug D. (1997) An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83, 85–90 10.1046/j.1365-2672.1997.00193.x [DOI] [PubMed] [Google Scholar]
- 31. Ostergaard Breum S., Neve H., Heller K. J., and Vogensen F. K. (2007) Temperate phages TP901–1 and ϕLC3, belonging to the P335 species, apparently use different pathways for DNA injection in Lactococcus lactis subsp. cremoris 3107. FEMS Microbiol. Lett. 276, 156–164 10.1111/j.1574-6968.2007.00928.x [DOI] [PubMed] [Google Scholar]
- 32. Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 34. Sonnhammer E. L., von Heijne G., and Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 35. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., et al. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 10.2144/03342mt01 [DOI] [PubMed] [Google Scholar]
- 37. McGrath S., Fitzgerald G. F., and van Sinderen D. (2001) Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67, 608–616 10.1128/AEM.67.2.608-616.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stockdale S. R., Collins B., Spinelli S., Douillard F. P., Mahony J., Cambillau C., and van Sinderen D. (2015) Structure and assembly of TP901–1 virion unveiled by mutagenesis. PLoS ONE 10, e0131676 10.1371/journal.pone.0131676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Pijkeren J. P., and Britton R. A. (2012) High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40, e76 10.1093/nar/gks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Pijkeren J. P., Neoh K. M., Sirias D., Findley A. S., and Britton R. A. (2012) Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3, 209–217 10.4161/bioe.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oh J. H., and van Pijkeren J. P. (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42, e131 10.1093/nar/gku623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Driscoll J., Glynn F., Cahalane O., O'Connell-Motherway M., Fitzgerald G. F., and Van Sinderen D. (2004) Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system. Appl. Environ. Microbiol. 70, 5546–5556 10.1128/AEM.70.9.5546-5556.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ciucanu I., and Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 10.1016/0008-6215(84)85242-8 [DOI] [Google Scholar]
- 44. Read S. M., Currie G., and Bacic A. (1996) Analysis of the structural heterogeneity of laminarin by electrospray-ionisation-mass spectrometry. Carbohydr. Res. 281, 187–201 10.1016/0008-6215(95)00350-9 [DOI] [PubMed] [Google Scholar]
- 45. Lindberg B., and Lönngren J. (1978) Methylation analysis of complex carbohydrates: general procedure and application for sequence analysis. Methods Enzymol. 50, 3–33 10.1016/0076-6879(78)50003-7 [DOI] [PubMed] [Google Scholar]
- 46. Kuipers O. P., de Ruyter P. G. G. A., Kleerebezem M., and de Vos W. M. (1998) Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21 10.1016/S0168-1656(98)00100-X [DOI] [Google Scholar]
- 47. Braun V., Hertwig S., Neve H., Geis A., and Teuber M. (1989) Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. Microbiology 135, 2551–2560 10.1099/00221287-135-9-2551 [DOI] [Google Scholar]
- 48. Higgins D. L., Sanozky-Dawes R. B., and Klaenhammer T. R. (1988) Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J. Bacteriol. 170, 3435–3442 10.1128/JB.170.8.3435-3442.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chandry P. S., Moore S. C., Boyce J. D., Davidson B. E., and Hillier A. J. (1997) Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26, 49–64 10.1046/j.1365-2958.1997.5491926.x [DOI] [PubMed] [Google Scholar]
- 50. Lillehaug D., Lindqvist B., and Birkeland N. K. (1991) Characterization of phiLC3, a Lactococcus lactis subsp. cremoris temperature bacteriophage with cohesive single-stranded DNA ends. Appl. Environ. Microbiol. 57, 3206–3211 10.1128/AEM.57.11.3206-3211.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oliveira J., Mahony J., Hanemaaijer L., Kouwen T. R. H. M., and van Sinderen D. (2018) Biodiversity of bacteriophages infecting Lactococcus lactis starter cultures. J. Dairy Sci. 101, 96–105 10.3168/jds.2017-13403 [DOI] [PubMed] [Google Scholar]
- 52. Oliveira J., Mahony J., Lugli G. A., Hanemaaijer L., Kouwen T., Ventura M., and van Sinderen D. (2016) Genome sequences of eight prophages isolated from Lactococcus lactis dairy strains. Genome Announc. 4, e00906–16 10.1128/genomeA.00906-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oliveira J., Mahony J., Hanemaaijer L., Kouwen T. R. H. M., Neve H., MacSharry J., and van Sinderen D. (2017) Detecting Lactococcus lactis prophages by mitomycin C-mediated induction coupled to flow cytometry analysis. Front. Microbiol. 8, 1343 10.3389/fmicb.2017.01343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rojas-Macias M. A., Mariethoz J., Andersson P., Jin C., Venkatakrishnan V., Aoki N. P., Shinmachi D., Ashwood C., Madunic K., Zhang T., Miller R. L., Horlacher O., Struwe W. B., Watanabe Y., Okuda S., et al. (2019) Towards a standardized bioinformatics infrastructure for N- and O-glycomics. Nat. Commun. 10, 3275 10.1038/s41467-019-11131-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data created, with the exception of MS data, can be found within the document. MS data have been deposited and are available on the Glycopost depository (54) with the ID GPST000056.