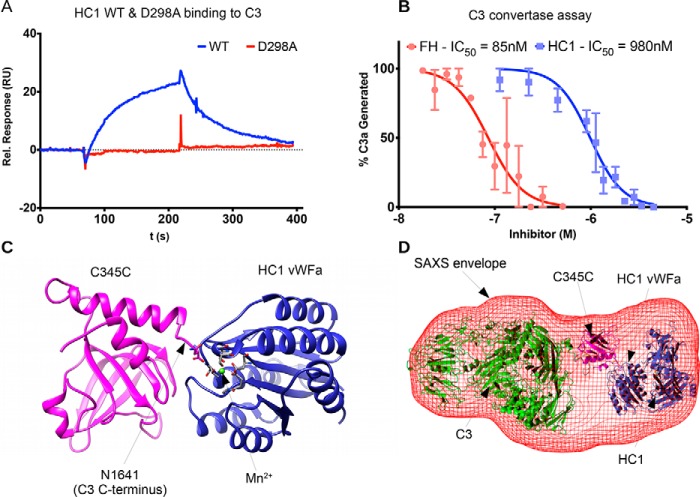
Figure 5.
HC1 inhibits alternative pathway C3 convertase activity through interaction with C3. A, SPR analysis for the interaction of rHC1 (WT and D298A) with C3 in 2 mm MnCl2, where the lack of binding of the D298A mutant indicates an essential role for the MIDAS site. B, rHC1 proteins (WT (blue) and D298A) were compared with FH (red) in an alternative pathway C3 convertase assay, where the proteolytic release of C3a was quantified (by SDS-PAGE) as a surrogate for the conversion of C3 into C3b. The mean values (± S.D.) were derived from independent experiments performed in triplicate. The data were fitted using GraphPad Prism to derive IC50 values for rHC1 and FH control. Only WT rHC1 had inhibitory activity (data for D298A not shown). C, an in silico model of the C3 C-terminal C345C domain (pink) bound to the vWFA domain of HC1 (blue). Here a Mn2+ ion (green) occupies the MIDAS of HC1 (with coordinating residues shown in stick representation) and co-chelates the C-terminal amino acid (Asn) of C3b. D, an ab initio SAXS structure was determined for the rHC1–C3 complex (red mesh), where C3 and HC1 molecules, interacting as in C, could be accommodated.