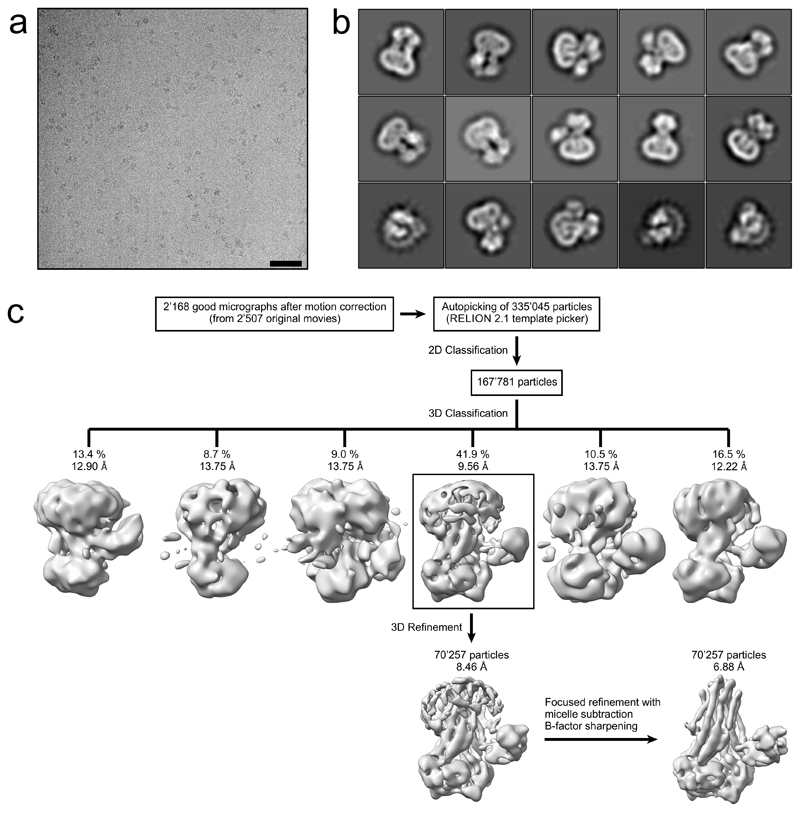
Extended Data Figure 2. Cryo-EM data processing of IrtAB.
a, Typical motion corrected cryo-EM micrograph of the detergent solubilized IrtAB sample. Scale bar: 50 nm. b, Representative 2D class averages used for 3D reconstruction, showing side and bottom views. c, Data processing workflow. 2’168 high-quality micrographs were selected after motion correction and used for autopicking of 335’045 particles. Several rounds of 2D classification were performed to remove false-positives, yielding 167’781 IrtAB particles, which were subjected to 3D classification. One out of six classes (indicated by black box), containing 70’257 particles, revealed distinct structural features of IrtAB with its SID and was subjected to 3D refinement. The resulting structure was used to subtract the detergent micelle from the raw particles. Afterwards, a focused 3D refinement without the detergent micelle was performed. The resulting map was sharpened using a B-factor of -319 Å2. The final structure was resolved to 6.88 Å and reveals details of the TMDs, the NBDs and the SID.