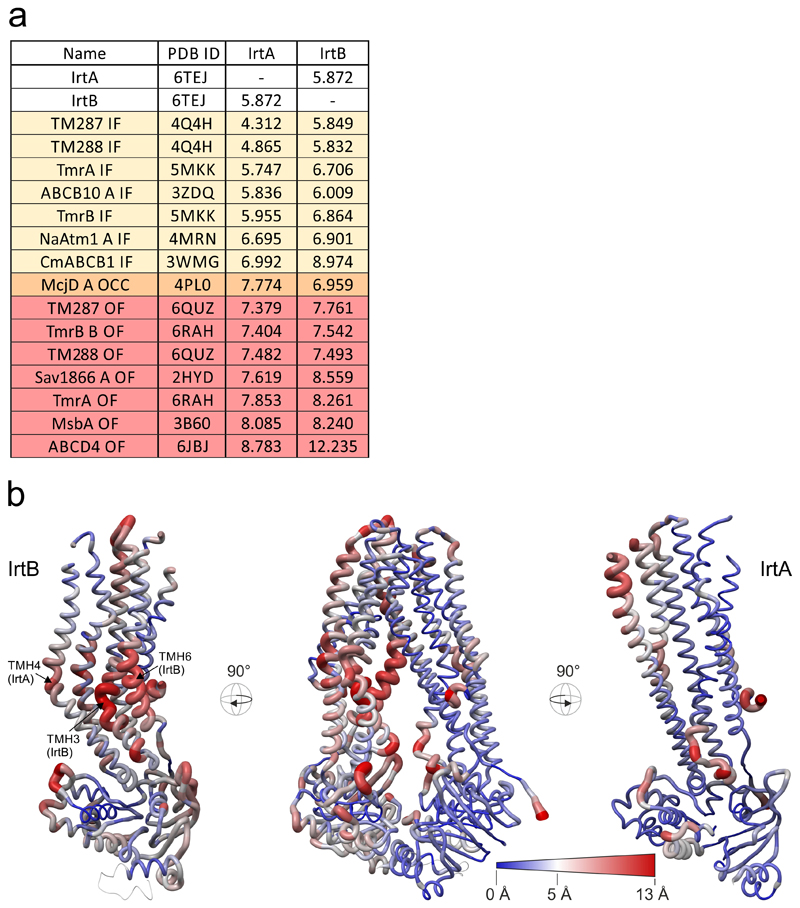
Extended Data Figure 4. Superimpositions with ABC exporter structures and structural comparison of IrtAB and TM287/288.
a, Superimposition of ABC exporters using the MatchMaker tool of Chimera. Nidelmann/Wunsch alignment algorithm and Blossom-62 matrix was used. R.m.s.d. (Å2) values were calculated over the entire polypeptide chains, i.e. without pruning of long atom pairs. In case the asymmetric unit contained several polypeptides of identical sequence, chain A was taken for analysis (indicated as “A” in the protein name). IF, inward-facing; OCC, occluded; OF, outward-facing. b, Structural deviations between IrtAB and the ABC exporter TM287/288 (PDB: 4Q4H). Shown is the structure of IrtAB in an “open book representation” with the entire transporter in the middle and the two half-transporters on the left (IrtB with domain-swap-helices TMH4 and 5 of IrtA) and on the right (IrtA with domain-swap-helices of IrtB). The structures are shown as sausage representation in which the thickness and color gradient from blue to red indicate variable degrees of structural deviations between IrtAB and TM287/288. Major structural deviations in the TMDs of IrtB are indicated.