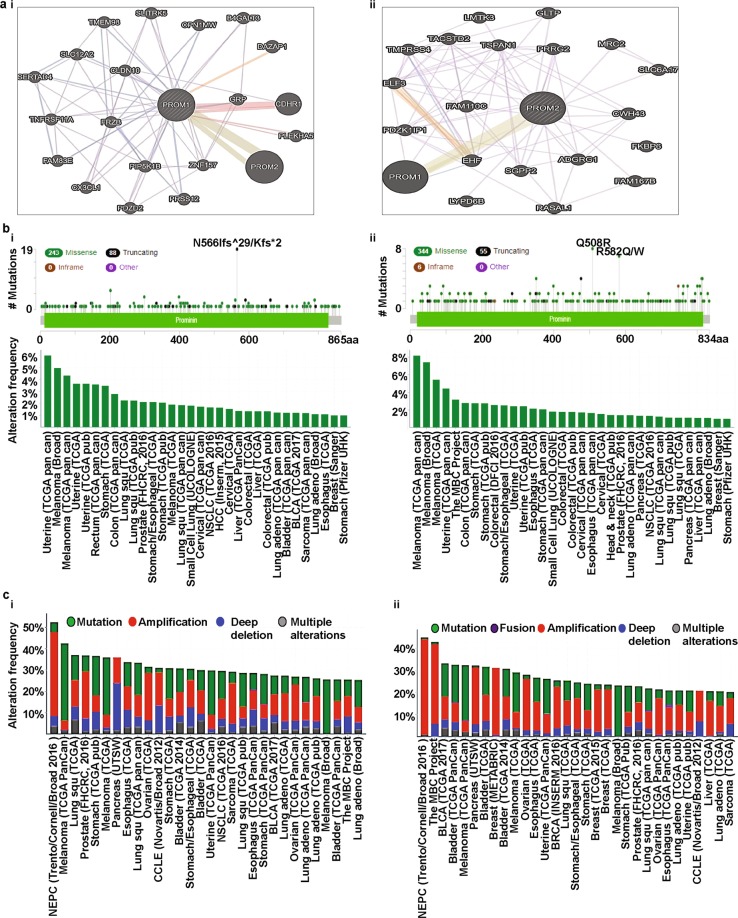
Fig. 5.
Identification of known and predicted structural proteins essential for PROM1 and PROM2 function (GeneMANIA) and frequency of mutations and copy number alterations (CNAs) in various types of cancer (cBioPortal web). a Interacting nodes are displayed in circles using GeneMANIA. Predicted functional partners of PROM1 (i) and PROM2 (ii) are shown after considering co-expression, co-localization, genetic interactions, pathways, physical interactions, and predicted shared protein domains. b In total, 331 mutation sites were identified and were located between amino acids 0 and 865 of PROM1. PROM1 mutations mainly occurred in uterine cancer and existed in a hotspot in the prominin domain (i). In total, 405 mutation sites were detected and were located between amino acids 0 and 834 of PROM2. PROM2 mutations mainly occurred in melanoma and also existed in a hotspot in the prominin domain (ii). c The alteration frequency of a ten-gene signature (PROM1, PROM2, GRP, ZNF157, FRZB, CLDN10, PIP5K1B, CDHR1, CX3CL1, and PDZD2) was determined using cBioPortal. Only data sets containing >100 samples per cancer type and an alteration frequency of >25% are shown. The alterations included mutations (green), amplifications (red), deep deletions (blue), or multiple alterations (grey) (i). The alteration frequency of a ten-gene signature (PROM2, PROM1, EHF, FAM110C, SGPP2, ADGRG1, CWH43, PRRG2, TSPAN1, and ELF3) was determined using cBioPortal. Only data sets containing >100 samples per cancer type and an alteration frequency of >20% are shown. The alteration frequency included mutations (green), fusions (brown), amplifications (red), deep deletions (blue), or multiple alterations (grey) (ii)