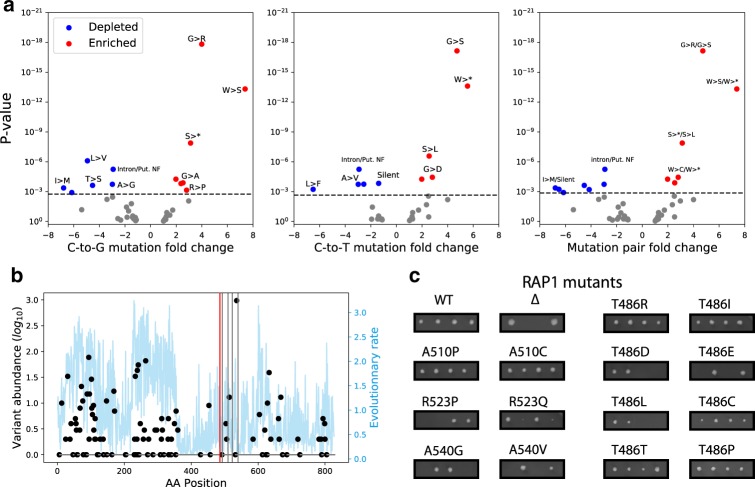
Fig. 5. GNE mutations are enriched for specific amino acid substitution patterns and identify critical sites for protein function.
a Fold depletion and enrichment volcano plots for the most probable mutations induced by GNEs in the screen. Enrichment and depletion values were calculated by comparing the relative abundance of each mutation among GNEs and NSGs using two-sided Fisher’s exact tests. Mutation patterns significantly depleted are shown in blue, while those that are enriched are in red. The significance threshold was set using the Holm–Bonferroni method at 5% FDR to correct for multiple testing and is shown as a dotted gray line. b Protein variant frequency among 1000 yeast isolates (black dots) and residue evolutionary rate across species (blue line) for RAP1. The target site for the GNEs targeting T486 is highlighted by a red line while the other detected GNEs target sites are shown by a gray line. c Tetrad dissections confirm most RAP1 GNE induced mutations indeed have strong fitness effects, as well as other substitutions targeting these sites. Source data are available in the Source Data file.