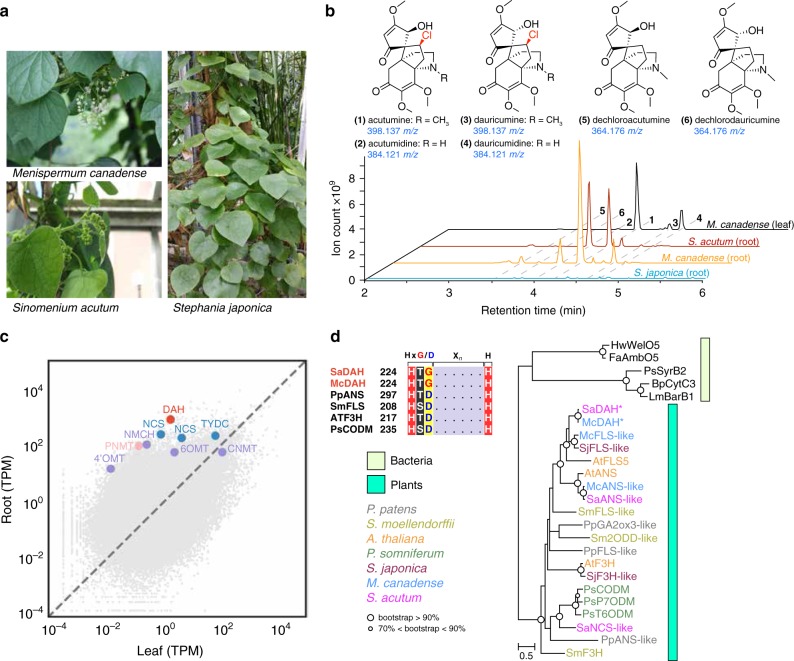
Fig. 1. Identification of candidate genes encoding DAH in Menispermaceae plants.
a Images of M. canadense, S. acutum, and S. japonica plants. b Extracted ion chromatograms (XICs) exhibiting the presence and absence of (−)-acutumine and its related compounds in three Menispermaceae plants. c Transcript differential expression analysis in M. canadense root vs. leaf tissues. Transcript abundance is quantified by transcripts per million (TPM) values. Candidate (−)-acutumine biosynthetic enzymes are colored based on their postulated roles in the proposed pathway depicted in Supplementary Fig. 1. d Sequence and phylogenetic analysis of DAH. The insert at the upper left corner shows multiple sequence alignment of two DAHs and other ODDs highlighting the Hx(D/G)XnH motif. The full sequence alignment is shown in Supplementary Fig. 3. The phylogenetic tree of DAHs together with select 2ODD-family proteins is inferred using maximum-likelihood method. Bootstrap statistics (200 replicates) are indicated at the tree nodes. The scale measures evolutionary distance in substitutions per amino acid. Asterisks denote the two orthologous DAHs identified in this study. The multiple sequence alignment used for building the phylogenetic tree is in Supplementary File 1. NCS norcoclaurine synthase, TYDC tyrosine decarboxylase, PNMT pavine N-methyltransferase, 6OMT (RS)-norcoclaurine 6-O-methyltransferase, CNMT (S)-coclaurine N-methyltransferase, NMCH (S)-N-methylcoclaurine-3-hydroxylase, 4′OMT (S)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase, ANS anthocyanidin synthase, FLS flavonol synthase, F3H flavanone-3-hydroxylase, CODM codeine O-demethylase, GA2ox3 gibberellin 2-beta-dioxygenase 3, 2ODD iron(II)- and 2-oxoglutarate-dependent dioxygenase, P7ODM papaverine 7-O-demethylase, T6ODM thebaine 6-O-demethylase.