Summary
Three-dimensional chromatin structures undergo dynamic reorganization during mammalian spermatogenesis; however, their impacts on gene regulation remain unclear. Here, we focused on understanding the structure-function regulation of meiotic chromosomes by Hi-C and other omics techniques in mouse spermatogenesis across five stages. Beyond confirming recent reports regarding changes in compartmentalization and reorganization of topologically associating domains (TADs), we further demonstrated that chromatin loops are present prior to and after, but not at, the pachytene stage. By integrating Hi-C and RNA-seq data, we showed that the switching of A/B compartments between spermatogenic stages is tightly associated with meiosis-specific mRNAs and piRNAs expression. Moreover, our ATAC-seq data indicated that chromatin accessibility per se is not responsible for the TAD and loop diminishment at pachytene. Additionally, our ChIP-seq data demonstrated that CTCF and cohesin remain bound at TAD boundary regions throughout meiosis, suggesting that dynamic reorganization of TADs does not require CTCF and cohesin clearance.
Subject Areas: Male Reproductive Endocrinology, Genomics, Transcriptomics
Graphical Abstract

Highlights
-
•
Chromatin loops are reorganized during mouse spermatogenesis, being absent in pacSC
-
•
CTCF and cohesin remain bound to pacSC chromatin while TADs and loops are lost
-
•
Chromatin accessibility per se is not involved in the loss of TADs or loops in pacSC
-
•
A/B compartments switching is related to meiosis-specific mRNAs and piRNAs expression
Male Reproductive Endocrinology; Genomics; Transcriptomics
Introduction
The process of chromatin organization in three-dimensions (3D) has been largely mysterious since the first recognition of chromatin structures (Dounce et al., 1972). Hi-C technology (high-throughput genome-wide chromatin conformation capture, Lieberman-Aiden et al., 2009) is developed from chromosome conformation capture (3C) (Dekker et al., 2002), which enables the global characterization of the 3D chromatin architectures. Results from Hi-C (original version and its derived versions such as in situ Hi-C) revealed that chromatin architecture comprises a hierarchy of structures in mammals, with chromatin A/B compartments at multi-megabase scale, topologically associating domains (TADs) at hundreds of kilobases scale, and chromatin loops at kilobases to hundreds of kilobases scale (Dixon et al., 2012, Lieberman-Aiden et al., 2009, Rao et al., 2014). Previous studies have indicated that TADs (Flavahan et al., 2016, Hnisz et al., 2016, Lupiáñez et al., 2015) and/or enhancer-promoter loops impact gene expression and cellular physiology (Bonev et al., 2017, Isoda et al., 2017, Weintraub et al., 2017) and transcription elongation also affects the 3D genome organization in specific regions (Heinz et al., 2018). However, the elimination of TADs and chromatin loops by rapid degradation of the cohesin complex only modestly affects gene transcription programs (Rao et al., 2017), and inhibiting transcription with α-Amanitin does not substantially affect the establishment of 3D chromatin organization during early development (Du et al., 2017, Ke et al., 2017). Thus, much remains to be learned about how differentially arranged 3D chromatin architectures control key biological processes.
Mammalian spermatogenesis is known to involve extensive chromatin re-organization, of which analyses using imaging techniques have revealed these dynamic changes (Hao et al., 2019, Sassone-Corsi, 2002). During spermatogenesis, a small number of primitive type A spermatogonia (priSG-A) differentiate into more developed spermatogonia including type A spermatogonia (SG-A) and type B spermatogonia (SG-B) in sequence via mitosis (de Rooij, 2001). Subsequently, meiosis is initiated and marked by double-strand break (DSB) formation and is followed by full synapses of homologous chromosomes in pachytene spermatocytes (pacSC) (Tong and Lin, 2018). After meiosis I, spermatocytes rapidly divide in the ensuing meiosis II stage to form haploid round spermatids (rST) and spermatozoa (SZ) via spermiogenesis. Recent efforts using omics technologies revealed dynamic changes in transcriptomes, DNA methylomes, chromatin accessibility, etc., during spermatogenesis (Gan et al., 2013, Hammoud et al., 2014, Helsel et al., 2017, Hermann et al., 2018, Law et al., 2019, Maezawa et al., 2018, Rathke et al., 2014, Sohni et al., 2019). These findings have motivated researches into understanding the detailed molecular events in spermatogenesis, such as the 3D chromatin architectural dynamics and its effects on transcription regulation.
Because spermatogenesis represents a fundamental biological process that involves highly orchestrated rearrangement of chromosomes, characterization of 3D chromatin architectures during meiosis and spermatogenesis has been a highly active research area. For example, 3D genome structures of mammalian spermatogenesis have recently been studied by four groups at different sub-stages in mouse and rhesus monkey (Alavattam et al., 2019, Patel et al., 2019, Vara et al., 2019, Wang et al., 2019). One common insight from these reports is that TADs are depleted and compartment integrity is relatively weak during the pachytene stage. At the compartment level, however, a recent study has identified compartments that are more refined in resolution than the conventional A and B compartments and these “refined compartments” alternate between transcribing and non-transcribing regions during spermatogenesis of rhesus monkeys (Wang et al., 2019). In addition, cohesin occupancy in active compartments was shown to affect expression in pacSC and rST (Vara et al., 2019). Despite these findings, the relationship between the 3D genome and chromatin accessibility and their contribution to meiosis-specific gene expression programs (e.g., piRNAs) in spermatogenesis remain unclear.
Here, by using Hi-C and a series of other complementary technologies including assay for transposase-accessible chromatin using sequencing (ATAC-seq), chromatin immunoprecipitation sequencing (ChIP-seq), and RNA sequencing (RNA-seq), we demonstrated the following advances provided by this work that were not already described in any of the earlier publications. First, chromatin loops are present prior to and after, but not at, the pachytene stage. Second, chromatin accessibility per se is not involved in the diminishment of TADs or chromatin loops at the pacSC stage. Third, although the spermatozoa (SZ) has similar 3D chromatin structures as the primary spermatogonia (priSG-A) at the level of TADs and compartments, these two stages of cells have distinct chromatin loops. Finally, by tracing several factors (meiosis, DSBs, piRNAs, etc.) during the reprogramming of 3D chromatin architectures, our study elucidated the influence of 3D architecture on these factors related to spermatogenesis and suggests that reprogrammed chromatin compartments and loops in spermatogenesis underline differential transcriptional regulations that support and maintain spermatogenesis and prepare the sperms for the subsequent possible embryogenesis.
Results
Higher-Order Chromatin Structures Are Reorganized during Mouse Spermatogenesis
To investigate the relationship between 3D genome structures and transcriptional regulation during mammalian spermatogenesis, we isolated spermatogenic cells from five different stages of mouse spermatogenesis (priSG-A, SG-A, pacSC, rST, and SZ) (Figure 1A) using the unit gravity sedimentation procedure (STA-PUT method) (Bellvé et al., 1977, Bryant et al., 2013, Gan et al., 2013, Hur et al., 2016, Korhonen et al., 2015, Liu et al., 2015, Luense et al., 2016, Wang et al., 2019) except for the SZ (see Transparent Methods). After validating the purities of these isolated cells (Figure S1A) by two methods, morphological characterization (Figure 1A) and immunofluorescence staining with cell-specific markers (Figure S1B), we performed in situ Hi-C with these different spermatogenic cells, respectively. The correlation analyses of the Hi-C data supported the high quality of our datasets among the biological replicates (Table S1 and Figure S2A), and we randomly sampled the same amount of Hi-C valid pairs (172,252,595) from spermatogenic cells of each stage to do the following analysis.
Figure 1.
Reprogramming of Compartments and Topologically Associating Domains (TADs) throughout Mouse Spermatogenesis
(A) The schematic of mouse spermatogenesis (top panel). Phase-contrast microscopic images show the morphological characterization of five isolated spermatogenic cell types, including primitive type A spermatogonia (priSG-A), type A spermatogonia (SG-A), pachytene spermatocytes (pacSC), round spermatids (rST), and spermatozoa (SZ). Scale bar, 50 μm.
(B) Heatmaps showing the normalized Hi-C interaction frequencies (ratio of observed interaction frequencies to expected interaction frequencies [observed/expected]) (100-kb bins, chromosome 1) (pooled data from two biological replicates).
(C) Heatmaps showing Pearson's correlation for Hi-C interactions (50 kb bins, chromosome 1), which captured genomic A/B compartmentalization patterns in each tested spermatogenic stage. PCA1 (the first eigenvalues) is shown underneath the heatmap and normalized transcriptomic coverage is shown at the bottom.
(D) Heatmaps showing averaged normalized Hi-C interaction frequencies (ratio of observed interaction frequencies to expected interaction frequencies [observed/expected]) of each tested spermatogenic stage around the TAD boundaries (which were defined based on identifications in priSG-A samples).
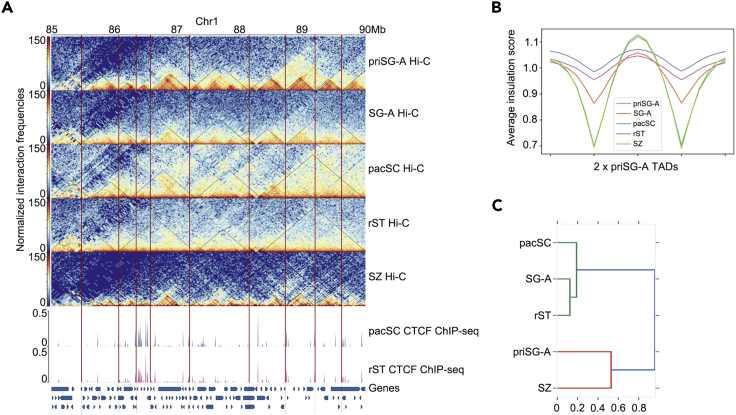
The chromatin interaction heatmaps revealed dramatic architectural reorganization throughout mouse spermatogenesis, as represented by chromosome 1 (Figure 1B), which confirmed recent reports using sub-stages (Vara et al., 2019, Wang et al., 2019). The A/B compartment patterns on autosomes were largely unaltered during mouse spermatogenesis (Figure 1C), whereas the strength, shown by saddle plots (Imakaev et al., 2012) (Figure S2B) and compartment strength (A-A and B-B compartments interaction strength relative to A-B compartments interaction strength) (Figure S2C) varied throughout the process. The strength of compartmentalization decreased from priSG-A to SG-A and dropped to the lowest level in pacSC before again increasing in rST and finally reaching its highest detected level in SZ. We also observed that the distinct TAD structures in priSG-A and SG-A largely disappeared in pacSC and rST. Interestingly, they were largely restored in the SZ stage (Figures 1D and 2A). This trend of TAD reorganization was most clearly manifested by global analysis of averaged observed/expected interaction frequencies centered on these boundaries (Figure 1D) and was also very apparent as seen on a random representative genomic region such as Chromosome 1, 85–90 Mb region (Figure 2A). We further adopted an “insulation score (IS)” approach to calculate the strength of TAD boundaries, which analyzes the relative chromatin interaction frequency across a boundary (Crane et al., 2015). We found that the IS values decreased gradually from priSG-A to pacSC, then increased from rST to SZ again, and the IS value of SZ almost exactly matched that of priSG-A (Figure 2B).
Figure 2.
Topologically Associating Domains (TADs) Are Reorganized during Mouse Spermatogenesis
(A) Chromatin observed/expected interaction frequencies views (25 kb bin) of cells at different tested spermatogenic stages. Black triangular lines marked TADs of each stage and red vertical lines indicated boundary loci at the priSG-A stage. CTCF ChIP-seq coverage at the pacSC and rST stages and gencode genes were depicted at the bottom.
(B) The average insulation scores (ISs) of cells at different spermatogenic stages at TADs (defined in the priSG-A stage) and nearby regions (±0.5 TADs length).
(C) Dendrogram showing the hierarchical clustering of IS values calculated with window size of 100 kb from interaction matrices (25 kb bin) of cells at different spermatogenic stages.
priSG-A and SZ Share Similar 3D Chromatin Interactions and Folding Patterns
With these IS values, we performed hierarchical clustering and found that TAD structures that once existed in priSG-A, and disappeared in pacSC, had reappeared in SZ, making these two stages (priSG-A and SZ) clustered together (Figure 2C). These data led us to examine the 3D genome structures at the different spermatogenic stages in more detail. We found that the global 3D chromatin interactions were similar between priSG-A, SG-A, and SZ (Figures 3A–3C), whereas pacSC showed stronger short-range chromosome interactions (<5 Mb) but weaker long-range chromosome interactions (>5 Mb) as compared with SG-A (Figure 3A). Specifically, the contact frequency P(s) with genomic distances demonstrated that 3D genome structures were reorganized during spermatogenesis (Figure 3C), showing that priSG-A, SG-A, and SZ had similar chromatin folding patterns and that pacSC represented an extreme case, consistent with the above IS hierarchical clustering data.
Figure 3.
Global 3D Architecture Reprograms throughout Mouse Spermatogenesis
(A) Heatmaps of ratio comparisons of the normalized Hi-C interaction frequencies (observed/expected) (100 kb bins, chromosome 1) at different tested spermatogenic stages.
(B) Heatmaps of ratio comparisons of the observed/expected interaction frequencies (100 kb bins, chromosome 1) for pacSC versus priSG-A, SZ versus pacSC, SZ versus priSG-A cells, respectively.
(C) The P(s) curves (relationship between interaction probability and genomic distance) for each tested spermatogenic stage.
(D) The P(s) curves (relationship between interaction probability and genomic distance) for pacSC and mitosis, and slopes (k) −1 (blue) and −0.5 (orange) were shown by the dotted lines.
Recent studies showed that mitotic chromatin exhibits a folding pattern distinct from that of interphase chromatin (Gibcus et al., 2018, Naumova et al., 2013). Because both meiotic and mitotic cells undergo chromosome condensation, we asked what is the difference between meiosis (pacSC) and mitosis in terms of 3D chromatin structures. The P(s) curves showed that at a scale less than 1 Mb, mitotic and meiotic chromatin shared similar folding patterns with a P(s)∼s slope at about −0.5 (Figure 3D). However, they displayed a striking difference at longer distances, where the meiotic P(s) curve had a steep drop at ∼8 Mb, whereas the mitotic curve dropped at ∼30 Mb (Figure 3D). This result suggested that meiotic pacSC chromatin is condensed via distinctive mechanisms as compared with that of mitotic chromatin.
Active Compartments for Genes and piRNA Clusters with Meiosis-Related Functions Are Enriched in pacSC
We further examined A/B compartmentalization in these spermatogenic stages in detail. The A compartments are defined as regions with a positive first eigenvector value when eigenvector decomposition is applied on observed/expected intra-chromatin interaction matrices (50 kb bin size in this study). These computed A compartments correspond to the active, euchromatic nuclear compartments. By contrast, the B compartments largely correspond to those inactive, heterochromatic genomic regions (Lieberman-Aiden et al., 2009). We investigated if there is A/B compartment switching between priSG-A and pacSC. We found A compartments accounted for 43.2% in priSG-A and 13.9% of them changed to B compartments in pacSC. Of the other 56.8% B compartments in priSG-A, 15.3% changed to A compartments in pacSC (Figure 4A). Notably, the number of genome regions in the A (active) compartments was higher in pacSC, suggesting that pacSC chromatin is in a more transcriptionally active state.
Figure 4.
Spermatogenesis-Related Genes and piRNA Clusters Are Located in Active Compartments Emerging in pacSC Cells
(A) Schematic of the proportion of genome regions that switched their compartment states (A to B or B to A) comparing between priSG-A and pacSC.
(B) Bar chart showing the numbers of differentially expressed genes located in compartments transiting between the A and B states.
(C) PCA1 (the first eigenvalues) and RNA-seq coverage track of chromosome 1, 53–58 Mb showed a locus where a given gene, Boll, exhibited differential expression at two different stages. PCA1 was calculated via eigenvector decomposition on the observed/expected intra-chromatin interaction matrices. Normalized values for the extent of transcriptomic coverage in priSG-A and pacSC were shown in the middle. Gencode genes were shown at the bottom.
(D) Bar chart showing the number of piRNA clusters positioned in A or in B compartments in cells at the priSG-A stage, and the number of piRNA clusters that switched to the other compartment state (A to B, or vice versa) in cells at the pacSC stage.
(E) Averaged ucsc phastCons scores on piRNA clusters stable within the A compartments at both the priSG-A and pacSC stages or on piRNA clusters that switched from the B to A compartment.
We then identified the genes harbored by compartments that showed switching behaviors and examined their expression using RNA-seq data at the corresponding priSG-A and pacSC stages (Lin et al., 2016). Although there were genes that switched from A to B or B to A compartments with both up- and down-regulation of their expression, we found that genes that were originally located in compartment B regions in priSG-A but switched to compartment A regions in pacSC, and at same time increased their expression, were the most abundant (Figure 4B, 1,037 genes). This result implies that compartment switching from B to A is correlated with up-regulation of gene expression for their proper biological functions. Indeed, a number of these genes function in cilium formation and DNA DSBs repair (Figure S3A), which are critical for normal sperm physiology and function (Figure S3B). One example for both compartments switching and increased gene expression in pachytene cells is the DAZ family protein, Boll (Figure 4C), which is required for the meiotic G2/M transition and germ cell development (VanGompel and Xu, 2010).
PIWI proteins and PIWI-interacting RNAs (piRNAs) direct the silencing of target nucleic acids in animal germline and soma (Ozata et al., 2019). In early pachytene, piRNAs are produced from large genomic loci referred to as piRNA clusters; these RNAs function in controlling transposons and have been shown to contribute to paternal imprinting (Aravin et al., 2007, Watanabe et al., 2011) and are required for the subsequent meiosis process (Fu and Wang, 2014). In addition to identifying genes in the compartments switched regions, we also analyzed the expression of piRNA clusters that locate to compartments showing stage-specific switching (piRNA clusters were identified from a published database, Rosenkranz, 2016). We found that 79.9% of all piRNA clusters had compartment A status in priSG-A, of which 88% remained in A compartments in pacSC; in contrast, 80% of the piRNA clusters with compartment B status in priSG-A switched to compartment A status in pacSC, correlating with their increased expression changes (Figure 4D).
To explore the difference between piRNA clusters that had compartment-A status in both priSG-A and pacSC cells (not switched) and that switched compartments from B to A between the priSG-A and pacSC stages, we examined the conservation status of these two categories of piRNA clusters using phastCons score, which measures the probability that each nucleotide belongs to a conserved element (Siepel et al., 2005). We averaged phastCons scores of every nucleotide in each piRNA cluster and compared the average scores of not switched and switched piRNA clusters between the priSG-A and pacSC stages. The former (not switched) was more conserved than the switched between these two stages in a phylogenetic analysis of 60 vertebrate species (Figure 4E). This result indicated that the piRNA clusters with compartment A status in both priSG-A and pacSC cells exhibit signs of selection pressure and are conserved among different species, suggesting functional constraints on them and thus their potential functions in these species. These data suggested that the functional activity of piRNAs might be strongly impacted by their locations in compartment A or B in the 3D genome.
Chromatin Accessibility and CTCF/Cohesin Binding Are Retained throughout Mouse Spermatogenesis
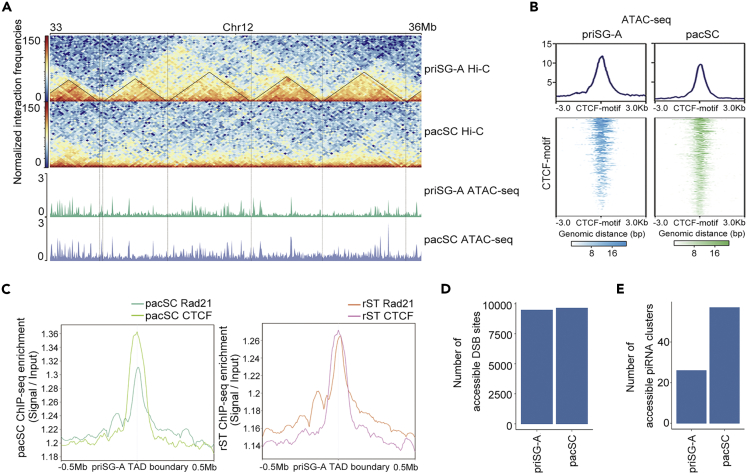
Since TADs dramatically changed throughout spermatogenesis and disappeared in pacSC cells, we asked whether the 3D architecture of pacSC cells correlates with other genomic/epigenomic features. We conducted ATAC-seq to measure chromatin accessibility (Buenrostro et al., 2015) in four spermatogenic cell types (priSG-A, SG-A, pacSC, and rST) (Figure S4A). We first found that the accessible chromatin regions were mostly located at gene promoters or enhancers and the intensity of the accessible chromatin signal decreased as spermatogenesis proceeds (Figures S4B and S4C). When we compared the chromatin accessibility in priSG-A and pacSC, we found minimal differences between them (Figure 5A). This similarity of chromatin accessibility was rather surprising because the 3D genome architecture in these two cell types was largely different (Figure 5A). These data suggested that 3D genome reorganization might not be functionally linked to chromatin opening, at least at the level of TADs.
Figure 5.
Genome Features of Accessible Regions at the pacSC Stage
(A) Chromatin observed/expected interaction frequencies views (25 kb bin) of cells at the priSG-A and pacSC stages, respectively. Vertical dashed lines indicated TAD boundary loci in priSG-A samples. Normalized ATAC-seq coverage in pacSC and rST samples were depicted at the bottom.
(B) Heatmaps showing the ATAC-seq signals enrichment around CTCF motif regions (±3kb), average ATAC-seq signals around all CTCF motif regions were plotted at the top.
(C) Profile plot showing the average CTCF and Rad21 signals (CTCF or Rad21 RPKM divided by input RPKM) in pacSC and rST samples around the TAD boundaries (±0.5Mb) defined in priSG-A samples.
(D) The number of accessible DSB sites in priSG-A and pacSC samples.
(E) The number of accessible piRNA clusters in priSG-A and pacSC samples.
Chromatin accessibility is closely related to transcriptional activity. We thus examined transcriptional activity using ChIP-seq with antibodies against the elongating form of RNA Polymerase II (Pol II S2P). The enrichment of Pol II S2P on gene transcriptional start sites (TSSs) showed that these genes were actively transcribing in both pacSC and rST (Figure S5A). However, the chromatin accessibility showed distinct patterns in pacSC and rST samples (Figure S5B). The chromatin accessibility in these two cell types was also very different from patterns in the other two cell types: priSG-A and SG-A (Figure S5B). Specifically, between pacSC and priSG-A, we identified 6,580 (out of 15,815) differentially accessible chromatin regions, whereas between pacSC and rST, there were 1,288 (of 15,815) differentially accessible regions. The chromatin regions displaying altered accessibilities between priSG-A and pacSC were analyzed for GO terms, and we found that they enriched for genes related to meiosis, and especially to the key events in meiosis I, e.g., meiotic recombination or synapsis (Figure S5C). A similar comparison of differentially accessible chromatin regions between pacSC and rST also revealed enrichment of meiotic genes, but notably, there was no longer any enrichment for gene groups specific to meiotic recombination or synapsis (Figure S5D). These results provided a molecular explanation of the biological knowledge that pacSC are at the prophase of the first meiotic division and that rST are produced after meiosis completion. Moreover, these data highlighted the utility of assessing chromatin accessibility states for inferring the functional status of transcriptional activity.
We then further investigated whether the differential chromatin accessibility may provide a molecular basis to the observed disappearance of TADs at the pacSC stage. As CTCF is enriched at TAD boundaries and the depletion of CTCF results in loss of TADs (Dixon et al., 2012, Nora et al., 2017), we checked if the chromatin accessibility of CTCF motifs may be affected at the TAD boundaries. Intriguingly, the chromatin accessibilities at CTCF motifs appeared highly similar between priSG-A and pacSC (Figure 5B), indicating that CTCF could bind to chromatin, which does not form conspicuous TADs (i.e., at the pacSC and rST stages). We experimentally examined CTCF binding in pacSC and rST cells via CTCF ChIP-seq. We found that, at both the pacSC stage and the rST stage, CTCF was still enriched at the TAD boundaries that were defined from the priSG-A stage (Figure 5C). In addition to CTCF, we also checked the binding of cohesin in pacSC and rST by Rad21 ChIP-seq. The result showed that in both pacSC and rST, Rad21 was also enriched at the TAD boundaries that were defined from the priSG-A stage (Figure 5C). These results suggested that the TAD disappearance in the meiotic stages of spermatogenesis could not be explained by the loss of CTCF or cohesin binding. Our data also renewed the understanding of CTCF and cohesin function in 3D genome structure, i.e., although CTCF and cohesin may be the molecules required for TAD maintenance in mammals (Nora et al., 2017, Rao et al., 2017), our data suggest that CTCF or cohesin binding at TAD boundaries is insufficient to maintain TADs at the pacSC stage.
During mammalian spermiogenesis, a highly condensed chromatin structure of sperm nuclei is in part formed by histone-protamine exchange and 1%–8% of histones in mouse are retained in sperm chromatin (Jung et al., 2019). Sperm histones were found to be mainly retained in distal intergenic regions, as shown by ChIP-seq using nucleoplasmin (NPM)-treated sperm (Yamaguchi et al., 2018). However, whether TAD boundaries play a role in retaining these histones in the sperm is unknown. We analyzed published protamine 1 targeted ChIP-seq data from sperms (Yoshida et al., 2018) and averaged the ChIP-seq signals around TAD boundaries in the sperm. Although protamine 1 was shown to occupy TSS regions, there was no enrichment of protamine 1 at the TAD boundaries (Figure S5E). We also analyzed the published histone H3 ChIP-seq data (Yamaguchi et al., 2018) and found no enriched localization of histone H3 around TAD boundaries, either (Figure S5F). These results implied that the TAD structure in the sperm may not be related to the histone or protamine constitution. Another question we asked was whether sperm-retained histones play substantial roles in the next generation. We found that, although histone H3 was depleted from the TSS regions, 171 genes had at least one exon in histone H3 retaining regions, and 16 of these 171 genes had been previously reported to be expressed in the early embryonic stage (Theiler stage 1–5, Table S2) (Smith et al., 2019). Further work examining the distribution of different modifications of retained histones and their potential roles in subsequent embryogenesis will provide more accurate answers to the function of retained histones.
Meiotic DSB Sites Are Pre-opened at the priSG-A Stage and piRNA Clusters Are Uniquely Open at the pacSC Stage
DNA double-strand break (DSB) is essential for the recombination in meiosis, and it begins at the leptotene stage and ends at the zygotene stage (Baudat et al., 2013, Smagulova et al., 2011). We examined the accessibility of chromatin regions surrounding previously reported DSB hotspots midpoints by analyzing our ATAC-seq data. We first identified meiotic DSB hotspots midpoints by the published Dmc1 ChIP-seq data obtained from DSB-positive cells in Hop2 knockout mice, whose meiotic DSBs are not repaired and meiotic progression is blocked at the pachytene-like stage (Smagulova et al., 2011). We then compared the chromatin accessibility around these meiotic DSB hotspots midpoints at the priSG-A and pacSC stages. Remarkably, this analysis revealed that the numbers of the accessible DSB hotspots midpoints were largely unchanged between these two stages (Figure 5D). These results raised an intriguing possibility that meiotic DSB hotspots might be pre-opened (i.e., "primed") prior to their starting point, perhaps at a time even earlier than the priSG-A stage, and remained open after their repairing at the pacSC stage.
Since we observed that there were more piRNA clusters located in active compartments at the pacSC stage than at the priSG-A stage (Figure 4D), we further compared the numbers of accessible pacSC piRNA clusters between priSG-A and pacSC. A larger number of accessible piRNA clusters were found in pacSC but not in priSG-A (Figure 5E). These data were consistent with the above observation that more piRNA clusters were located in active compartments at the pacSC stage compared with that at the priSG-A stage, and it was also in line with the fact that pacSC-specific piRNAs are transcribed at the pacSC stage. These data together supported a refined coordinated functional contribution of DSBs and piRNA clusters to the meiosis process of murine spermatogenesis.
Reorganized Chromatin Loops in Sperm Contribute to Early Embryo Development
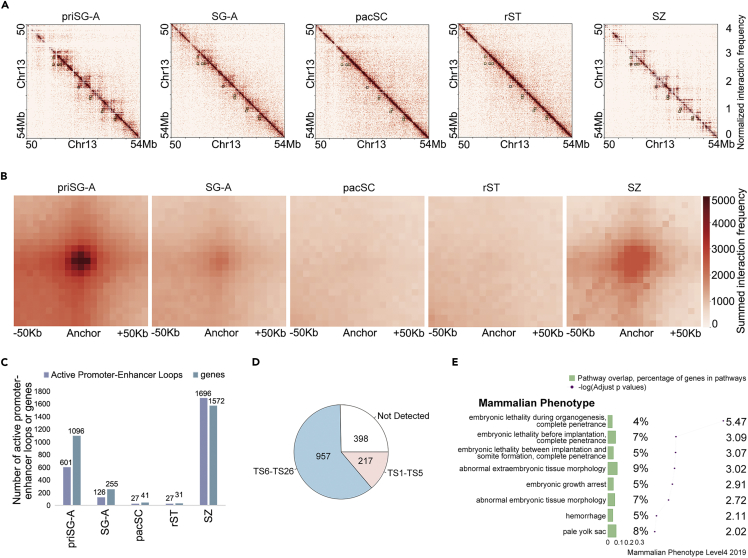
To examine the finer-scale architecture of the chromatin, we identified chromatin loops from the Hi-C data in each spermatogenic stage using cLoops (Cao et al., 2020). We observed that these loop interactions were reprogrammed throughout the spermatogenesis process (Figures 6A and 6B), similar to the dynamics at TAD or compartment strength levels. The number of loops was significantly decreased at the pacSC and rST stages, followed by an increase to a maximum value in SZ (Figure 6C). We aggregated the interaction values around loops defined in priSG-A and found that the strength of loops, as shown by the APA values (Rao et al., 2014), showed the same trend as the loop numbers (Figure S6A).
Figure 6.
Reorganization of Chromatin Loops during Mouse Spermatogenesis and Their Implicated Roles in Early Embryo
(A) Heatmaps showing the observed/expected Hi-C interaction frequencies (5 kb bin, chromosome 13, 50–54 Mb) of cells at different tested spermatogenic stages. Black boxes showed the loop loci that were defined in priSG-A samples.
(B) Heatmaps showing the summed obs/exp Hi-C interaction frequencies within the vicinity (±50 kb) of the loop centers defined in priSG-A samples.
(C) The number of active promoter-enhancer loops at different spermatogenic stages. Loops were defined by cLoops. Enhancers were marked by H3K4me1 or H3K27ac outside promoter regions in testis, and active promoters in testis were indicated by enrichment of H3K4me3 or Pol II binding signals. The number of genes whose promoters were overlapped with loop anchors was shown.
(D) Pie plot showing the number of genes located in enhancer-promoter loops in SZ samples according to whether their expression was detected in early embryonic stages (Theiler stages 1–5) or in Theiler stages 6–26.
(E) GO-based mammalian phenotype enrichment data based on 217 genes positioned at enhancer-promoter loops expressed in early embryonic stages (Theiler stages 1–5), and a bar plot showing the proportion of these genes pertaining to the annotation of each phenotype over the number of all genes annotated in each phenotype. Line plot showing the significance (-log (adjusted p values)) of each ontology.
To test the functional roles of the most abundant loops formed in SZ, we used ChIP-seq data of histone marks and RNA Pol II binding in testis from previous reports (Shen et al., 2012) to annotate potential enhancers. This analysis revealed that 1,572 genes were putatively involved in the formation of promoter-enhancer loops at the SZ stage (Figure 6C). Of these 1,572 genes, only a small percentage (317, or 20%) were also anchored in loops in priSG-A samples (Figure S6B), suggesting that, although chromatin architecture in SZ samples restored the pattern in priSG-A at the larger scale (of compartments and TADs), at the smaller scale, the reestablished loops in SZ samples were distinct from those in the priSG-A samples. Genes that were involved in loops in SZ might tend to be transcriptionally active in subsequent embryonic stages, and previous studies have demonstrated the expression of a majority (1,174, or 75%) of these 1,572 genes in embryos, with 217 genes specifically detected in the developmental window of Theiler stages 1–5, although many of them repeatedly appeared in different stages of these early stages (Smith et al., 2019) (Figure 6D). Specifically, 63 genes could be detected from Theiler stage 2 and 26 genes could be detected from Theiler stage 1 until Theiler stage 4 (Figure S6C). These 217 genes were found to be involved in biological processes like regulation of gene expression and cell differentiation (Figure S6D), and their reported knockout phenotypes in mammals include embryonic growth abnormality, arrest, and lethality (Figure 6E). As examples, mice showed embryonic growth arrest (Vauti et al., 2007) and mice showed abnormal embryonic erythropoiesis and abnormal liver development (Wang et al., 2000). These results implied that the reorganized loops we detected in SZ might facilitate the expression of genes required for cell differentiation during the subsequent embryogenesis.
Discussion
Recent studies have shown the dynamic changes in TADs and chromatin compartmentalization during mammalian spermatogenesis (Alavattam et al., 2019, Patel et al., 2019, Vara et al., 2019, Wang et al., 2019). Here, we further explored dynamic higher-order chromatin structures and their functional impacts on transcriptional regulation during mouse spermatogenesis combining Hi-C, ATAC-seq, ChIP-seq, and RNA-seq data. Our results complement recent reports by demonstrating that chromatin loops are dynamically reorganized during spermatogenesis in a pattern similar to that of TADs: they are present at stages prior to pacSC, are “erased” in pacSC, and get re-established after it. We also showed that the switching of A/B compartments between spermatogenic stages is tightly associated with meiosis-specific mRNAs and piRNAs expression. For mechanism, we showed that chromatin accessibility and the binding of CTCF and cohesin at TAD boundaries are not responsible for the TAD and loop diminishment at the pachytene stage.
It was reported that A/B compartment switching is correlated with the expression of specific genes during spermatogenesis (Vara et al., 2019). We also found some meiosis-related genes switch A/B compartment states and change expression between priSG-A and pacSC. In addition, we focused on piRNA clusters and demonstrated that most piRNA clusters are located in active A compartments and their chromatin states are quite stable, yet a majority of the piRNA clusters that are located in inactive B compartments in priSG-A are switched to active A compartments in pacSC. This is in line with the fact that pacSC cells are known to transcribe a characteristic set of pacSC-specific piRNAs (Aravin et al., 2006, Girard et al., 2006, Ozata et al., 2019); thus, our findings indicate that the chromatin compartment switch likely affects the activity of piRNAs.
Our results showed that, although the 3D genome organization changes greatly at the pacSC stage, chromatin at regions containing meiotic genes are still accessible, indicating that chromatin accessibility per se is not functionally associated with TADs or chromatin loops at the pacSC stage. In addition, enrichment of meiotic DSB sites in active A compartments has been reported during the pachytene stage (Patel et al., 2019). Chromatin accessibility was suggested to correlate with the occurrence of meiotic DSBs by some authors (Patel et al., 2019), but others have shown that accessible chromatin regions in pacSC have limited overlap with DSB sites (Maezawa et al., 2018). In the present study, we showed that many meiotic DSB sites are in accessible chromatin regions, and our data unexpectedly revealed that these sites are already "primed" in the priSG-A stage, which may play a functional role in generating DSB sites.
Chromatin tracing by super-resolution imaging at the single-cell level revealed TAD-like structures and showed that TAD boundaries tend to be located near CTCF and cohesin binding sites (Bintu et al., 2018). These TAD-like structures were still found in individual cells after removal of cohesin from chromatin, although the signal for the TAD-like structures was buried when the cells were analyzed as a population (Bintu et al., 2018). Upon specific chemical degradation of cohesin, the boundaries of the TAD-like structures in single cells appeared to be randomly distributed, suggesting that cohesin might not be required for TAD maintenance but may rather function to establish developmentally appropriate TAD boundaries (Bintu et al., 2018). Moreover, cohesin-mediated transcription in genomic regions out of the chromosomal axes in primary spermatocytes can provide an environment conducive to both gene expression and the formation of DSBs (Vara et al., 2019). We speculate that the population-averaged signal for TADs in the CTCF-depleted cells of the previous report (Nora et al., 2017) might also mask the occupancy of TAD structures in individual cells. However, this is not the case for meiotic chromatin: our ChIP-seq data showed that specific binding of CTCF and cohesin (at TAD boundaries that were defined at the priSG-A stage) is retained at the pacSC stage when TADs disappear, suggesting that there are additional factors that regulate the formation or maintenance of TAD structures, at least during meiosis. Multiple methodological strategies that combine 3D genome detection and immunoprecipitation or chromatin isolation and mass spectrometry and experiments better control for variables could be employed to identify candidates of such factors. In addition, loss of TADs and CTCF binding was reported to occur in mitotic chromatin (Gibcus et al., 2018, Naumova et al., 2013, Oomen et al., 2019), which, when taken together with our findings, strongly implies that TADs maintenance may be controlled via distinct mechanisms in mitosis versus meiosis.
In addition to lost TADs and chromatin loops, the global chromatin organization between mitotic and meiotic chromatin is also similar when compared with that in interphase, as revealed by our P(s) curve analysis. Nevertheless, meiotic chromatin exhibits A/B compartments (Figure 1C), whereas mitotic chromatin does not (Gibcus et al., 2018). Thus, a comparison of factors modulating 3D genome organization at the compartmentalization level in meiotic and mitotic chromatin may discover unknown factors involved in higher-order chromatin organization. In addition to CTCF binding, transcriptional activity represents another major difference between meiosis and mitosis and is known to be affected by higher-order chromatin organization. Although both meiotic and mitotic chromatin are compacted, meiotic chromatin remains transcriptionally active (Figure S5A), whereas mitotic chromatin is largely transcriptionally inert (Gottesfeld and Forbes, 1997, Wang et al., 2019). We propose that the distinct chromatin interaction patterns may be one of the causes underlying this difference.
Previous studies have reported that TADs and chromatin loops are associated with gene expression (Bonev and Cavalli, 2016, Flavahan et al., 2016, Hnisz et al., 2016, Lupiáñez et al., 2015), whereas there are also some findings that show that loss of TADs and chromatin loops by rapid degradation of cohesin or CTCF have only modest effects on gene expression (Nora et al., 2017, Rao et al., 2017). Thus, the impacts of chromatin organization at TAD and chromatin loop scales on gene expression are still not very clear. A recent study by Micro-C that explores the chromatin organization from single nucleosomes to whole genome-scale found that there are microTADs and gene-level folding within TADs that form in a gene-dependent manner and are highly associated with chromatin accessibility and transcriptionally active chromatin (Hsieh et al., 2020). Here, our results together with several recent reports show that TADs and chromatin loops are almost lost at the pacSC stage during mouse spermatogenesis (Patel et al., 2019, Vara et al., 2019, Wang et al., 2019), whereas we also show that CTCF/cohesin binding on chromatin, chromatin accessibility at promoter/enhancer regions, and Pol II binding remain at the pacSC stage. Thus, although TADs and chromatin loops are largely lost at the pacSC stage, the microTADs and gene-level folding, which cannot be detected by in situ Hi-C, might remain on pacSC chromatin and contribute to transcription regulation and gene expression at the pacSC stage. Our chromatin accessibility and Pol II S2P ChIP-seq data support this possibility, and future studies with nucleosome-resolution Micro-C and single-cell methods will further deepen our understanding of the relationship between chromatin organization and transcription regulation during mouse spermatogenesis. Altogether, beyond confirming several major conclusions about TAD reorganization and the dynamics of A/B compartments strength, our study also reveals mechanistic insights about the functional relationships between 3D genome organization, CTCF or cohesin binding, and TAD maintenance and demonstrates the roles of A/B compartments switching in controlling meiotic gene transcription programs as well as potential function of chromatin loops in SZ in preparing for subsequent embryogenesis.
Limitations of the Study
In this study, we explored the 3D genome organization during mouse spermatogenesis with isolated spermatogenic cells from five different stages. We showed that chromatin loops and TADs are reorganized during mouse spermatogenesis, which almost disappear at the pacSC stage, whereas the transcriptional activity, CTCF binding on chromatin, and chromatin accessibility of promoters and enhancers are largely maintained at the pacSC stage. Thus, it is unclear how the transcriptional regulation is going on at the pacSC stage when the pacSC chromatin has no chromatin loops and TADs. A recent study about transcription-linked mammalian 3D chromatin folding with Micro-C uncovered a finer-scale genome organization that is associated with transcription and chromatin accessibility more tightly, compared with chromatin loops and TADs (Hsieh et al., 2020). Future studies that focus on the finer-scale genome organization at the pacSC stage might reveal the relationship between 3D genome structures and transcriptional regulation, as whether the finer-scale genome organization remains at the pacSC stage while chromatin loops and TADs are almost lost is still unclear.
In addition, we and others only explored the 3D genome organization during mouse and rhesus monkey spermatogenesis with limited isolated cell types (Alavattam et al., 2019, Patel et al., 2019, Vara et al., 2019, Wang et al., 2019), which restrict us to get a more comprehensive understanding about the 3D genome organization during mouse spermatogenesis. Future studies with single-cell Hi-C technologies and related methods might give us a more comprehensive view about the 3D genome organization during mouse spermatogenesis and uncover the underlying mechanisms about the reorganized chromatin structures.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We sincerely thank Dr. Mingxi Liu (Nanjing Medical University), Dr. Bo Wen (Fudan University), and Dr. Yong Zhang (Institute of Zoology, Chinese Academy of Sciences) for their helpful advices. We greatly appreciate Dr. Shouhong Guang (University of Science and Technology of China, USTC) for insightful editing of the manuscript. We gratefully acknowledge English language assistance from Prof. Zachary J. Smith (USTC). This work was supported by grants from the National Key Research and Development Program of China (No. 2018YFC1003500 to F.S.), National Key Scientific Program of China (No. 2016YFA0100502 and 2015CB943002 to X.S.), the National Natural Science Foundation of China (Grant No. 31671490 to X.S., 81671510 to F.S., and 81901528 to X.W.), and the Cancer Prevention and Research Institute of Texas (CPRIT) Scholar Award (RR160083 to W.L.).
Author Contributions
Z.L. and X.W. performed experiments with assistance from J.C. (under the guidance of C.H.), X.G. (under the guidance of J.W.) and Q.X.; H.J. and R.W. performed data analysis with assistance from Y.C. (under the guidance of Y.Y.) and J.C.; X.S. and F.S. designed the project; X.S., Z.L., H.J., R.W., X.W., C.H., W.L., C.Y.C., and M.G.R. wrote and reviewed of manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: April 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101034.
Contributor Information
Fei Sun, Email: sunfei@ntu.edu.cn.
Xiaoyuan Song, Email: songxy5@ustc.edu.cn.
Supplemental Information
References
- Alavattam K.G., Maezawa S., Sakashita A., Khoury H., Barski A., Kaplan N., Namekawa S.H. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat. Struct. Mol. Biol. 2019;26:175–184. doi: 10.1038/s41594-019-0189-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Baudat F., Imai Y., de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bellvé A.R., Cavicchia J., Millette C.F., O'brien D.A., Bhatnagar Y., Dym M. Spermatogenic cells of the prepuberal mouse: isolation and morphological characterization. J. Cel. Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B., Mateo L.J., Su J.-H., Sinnott-Armstrong N.A., Parker M., Kinrot S., Yamaya K., Boettiger A.N., Zhuang X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362:eaau1783. doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Cavalli G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016;17:661. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J.M., Meyer-Ficca M.L., Dang V.M., Berger S.L., Meyer R.G. Separation of spermatogenic cell types using STA-PUT velocity sedimentation. J. Vis. Exp. 2013;80:e50648. doi: 10.3791/50648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109(1):21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Chen Z., Chen X., Ai D., Chen G., McDermott J., Huang Y., Guo X., Han J.J. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics. 2020;36:666–675. doi: 10.1093/bioinformatics/btz651. [DOI] [PubMed] [Google Scholar]
- Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–U299. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D.G. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dounce A.L., Chanda S.K., Ickowicz R., Volkman D., Palermiti M., Turk R. The structure of eukaryote chromatin. Acta Endocrinol. Suppl. 1972;168:86–111. doi: 10.1530/acta.0.071s086. [DOI] [PubMed] [Google Scholar]
- Du Z., Zheng H., Huang B., Ma R., Wu J., Zhang X., He J., Xiang Y., Wang Q., Li Y. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suvà M.L., Bernstein B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Wang P.J. Mammalian piRNAs. Spermatogenesis. 2014;4:e27889. doi: 10.4161/spmg.27889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C.-x., Wang M., He C., Han C. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 2013;4:1995. doi: 10.1038/ncomms2995. [DOI] [PubMed] [Google Scholar]
- Gibcus J.H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J., Kanemaki M.T., Xie L., Paulson J.R., Earnshaw W.C. A pathway for mitotic chromosome formation. Science. 2018;359:eaao6135. doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J.M., Forbes D.J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Hammoud S.S., Low D.H., Yi C., Carrell D.T., Guccione E., Cairns B.R. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Hao S.-L., Ni F.-D., Yang W.-X. The dynamics and regulation of chromatin remodeling during spermiogenesis. Gene. 2019;706:201–210. doi: 10.1016/j.gene.2019.05.027. [DOI] [PubMed] [Google Scholar]
- Heinz S., Texari L., Hayes M.G.B., Urbanowski M., Chang M.W., Givarkes N., Rialdi A., White K.M., Albrecht R.A., Pache L. Transcription elongation can affect genome 3D structure. Cell. 2018;174:1522–1536.e22. doi: 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A.R., Yang Q.E., Oatley M.J., Lord T., Sablitzky F., Oatley J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernstroer B. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667.e8. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Day D.S., Young R.A. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Rando O.J., Tjian R. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S.K., Freschi A., Ideraabdullah F., Thorvaldsen J.L., Luense L.J., Weller A.H., Berger S.L., Cerrato F., Riccio A., Bartolomei M.S. Humanized H19/Igf2 locus reveals diverged imprinting mechanism between mouse and human and reflects Silver–Russell syndrome phenotypes. Proc. Natl. Acad. Sci. U S A. 2016;113:10938–10943. doi: 10.1073/pnas.1603066113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T., Moore A.J., He Z., Chandra V., Aida M., Denholtz M., Piet van Hamburg J., Fisch K.M., Chang A.N., Fahl S.P. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell. 2017;171:103–119.e18. doi: 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.H., Kremsky I., Gold H.B., Rowley M.J., Punyawai K., Buonanotte A., Lyu X., Bixler B.J., Chan A.W.S., Corces V.G. Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol. Cell. 2019;75:154–171.e5. doi: 10.1016/j.molcel.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y., Yao X., Li F., Zhu W., Gao L. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170:367–381.e20. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Korhonen H.M., Yadav R.P., Da Ros M., Chalmel F., Zimmermann C., Toppari J., Nef S., Kotaja N. DICER regulates the formation and maintenance of cell-cell junctions in the mouse seminiferous epithelium. Biol. Reprod. 2015;93:139. doi: 10.1095/biolreprod.115.131938. [DOI] [PubMed] [Google Scholar]
- Law N.C., Oatley M.J., Oatley J.M. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat. Commun. 2019;10:2787. doi: 10.1038/s41467-019-10596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., Van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Han M., Cheng L., Chen J., Zhang Z., Shen T., Wang M., Wen B., Ni T., Han C. Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol. 2016;13:1011–1024. doi: 10.1080/15476286.2016.1218588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Niu M., Yao C., Hai Y., Yuan Q., Liu Y., Guo Y., Li Z., He Z. Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci. Rep. 2015;5:8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luense L.J., Wang X., Schon S.B., Weller A.H., Shiao E.L., Bryant J.M., Bartolomei M.S., Coutifaris C., Garcia B.A., Berger S.L. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin. 2016;9:24. doi: 10.1186/s13072-016-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa S., Yukawa M., Alavattam K.G., Barski A., Namekawa S.H. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018;46:593–608. doi: 10.1093/nar/gkx1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N., Imakaev M., Fudenberg G., Zhan Y., Lajoie B.R., Mirny L.A., Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Goloborodko A., Valton A.-L., Gibcus J.H., Uebersohn A., Abdennur N., Dekker J., Mirny L.A., Bruneau B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen M.E., Hansen A.S., Liu Y., Darzacq X., Dekker J. CTCF sites display cell cycle–dependent dynamics in factor binding and nucleosome positioning. Genome Res. 2019;29:236–249. doi: 10.1101/gr.241547.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata D.M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Patel L., Kang R., Rosenberg S.C., Qiu Y., Raviram R., Chee S., Hu R., Ren B., Cole F., Corbett K.D. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol. 2019;26:164–174. doi: 10.1038/s41594-019-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C., Baarends W.M., Awe S., Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta. 2014;1839:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Rosenkranz D. piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res. 2016;44(D1):D223–D230. doi: 10.1093/nar/gkv1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I.V., Brick K., Khil P., Camerini-Otero R.D., Petukhova G.V. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M., Hayamizu T.F., Finger J.H., Bello S.M., McCright I.J., Xu J., Baldarelli R.M., Beal J.S., Campbell J., Corbani L.E. The mouse gene expression database (GXD): 2019 update. Nucleic Acids Res. 2019;47:D774–D779. doi: 10.1093/nar/gky922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohni A., Tan K., Song H.W., Burow D., de Rooij D.G., Laurent L., Hsieh T.C., Rabah R., Hammoud S.S., Vicini E. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 2019;26:1501–1517.e4. doi: 10.1016/j.celrep.2019.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M.H., Lin Z. m(6)A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta. 2018;1862:403–411. doi: 10.1016/j.bbagrm.2018.10.016. [DOI] [PubMed] [Google Scholar]
- VanGompel M.J., Xu E.Y. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum. Mol. Genet. 2010;19:2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara C., Paytuvi-Gallart A., Cuartero Y., Le Dily F., Garcia F., Salva-Castro J., Gomez H.L., Julia E., Moutinho C., Aiese Cigliano R. Three-dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep. 2019;28:352–367.e9. doi: 10.1016/j.celrep.2019.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauti F., Prochnow B.R., Freese E., Ramasamy S.K., Ruiz P., Arnold H.H. Arp3 is required during preimplantation development of the mouse embryo. FEBS Lett. 2007;581:5691–5697. doi: 10.1016/j.febslet.2007.11.031. [DOI] [PubMed] [Google Scholar]
- Wang Q., Khillan J., Gadue P., Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang H., Zhang Y., Du Z., Si W., Fan S., Qin D., Wang M., Duan Y., Li L. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol. Cell. 2019;73:547–561.e6. doi: 10.1016/j.molcel.2018.11.019. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Tomizawa S.-i., Mitsuya K., Totoki Y., Yamamoto Y., Kuramochi-Miyagawa S., Iida N., Hoki Y., Murphy P.J., Toyoda A. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A.S., Li C.H., Zamudio A.V., Sigova A.A., Hannett N.M., Day D.S., Abraham B.J., Cohen M.A., Nabet B., Buckley D.L. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588.e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Hada M., Fukuda Y., Inoue E., Makino Y., Katou Y., Shirahige K., Okada Y. Re-evaluating the localization of sperm-retained histones revealed the modification-dependent accumulation in specific genome regions. Cell Rep. 2018;23:3920–3932. doi: 10.1016/j.celrep.2018.05.094. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Muratani M., Araki H., Miura F., Suzuki T., Dohmae N., Katou Y., Shirahige K., Ito T., Ishii S. Mapping of histone-binding sites in histone replacement-completed spermatozoa. Nat. Commun. 2018;9:3885. doi: 10.1038/s41467-018-06243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.