FIG 2.
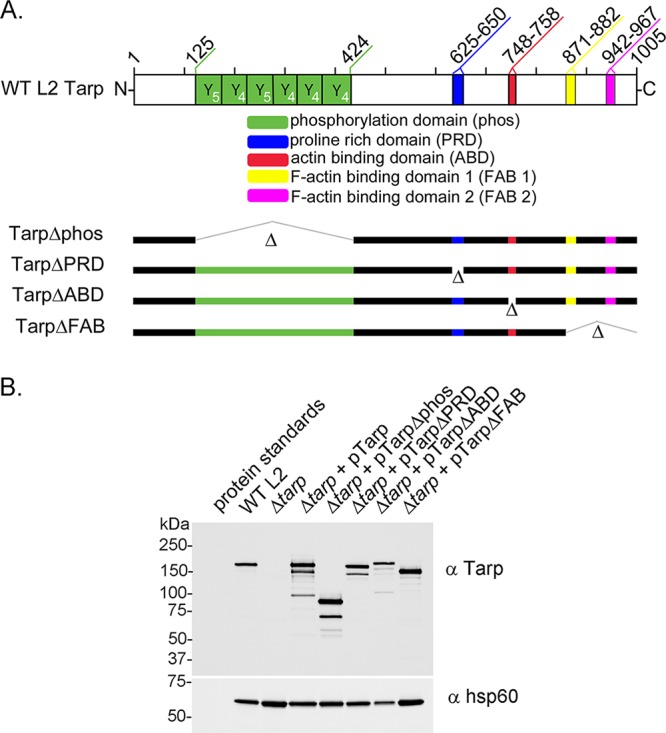
Schematic representation of Tarp and the complementation clones expressed in the tarP mutant. (A) Chlamydia trachomatis (L2) Tarp harbors an N-terminal tyrosine-rich repeat region, which is also referred to as the phosphorylation domain (phos; YYY, green boxes), a proline-rich domain (PRD; blue box), a single G-actin-binding domain (ABD; red box), and two F-actin-binding domains (yellow box [FAB 1] and pink box [FAB 2]). In-frame deletions engineered to remove specific protein domains for complementation are indicated with hatch marks and the corresponding Δ designation (Δphos, ΔPRD, ΔABD, ΔFAB 1, and ΔFAB 2). Numbers along the top of the schematic indicate amino acid positions encoded within the C. trachomatis L2 tarP gene. (B) Protein extracts from density gradient purified wild-type C. trachomatis L2 (WT L2), tarP deletion mutant (Δtarp), and mutants complemented from plasmid-based expression employing the Tarp promoter with full-length tarP (Δtarp + pTarp), tarP lacking the phosphorylation domain (Δtarp + pTarpΔphos), tarP lacking the proline-rich domain (Δtarp + pTarpΔPRD), tarP lacking the actin-binding domain (Δtarp + pTarpΔABD), or tarP lacking the F-actin-binding domains (Δtarp + pTarpΔFAB). EBs were resolved by SDS-PAGE and visualized by immunoblotting analysis with Tarp (α Tarp) and C. trachomatis heat shock protein 60 (α Hsp60)-specific antibodies. The molecular masses of protein standards are shown.
