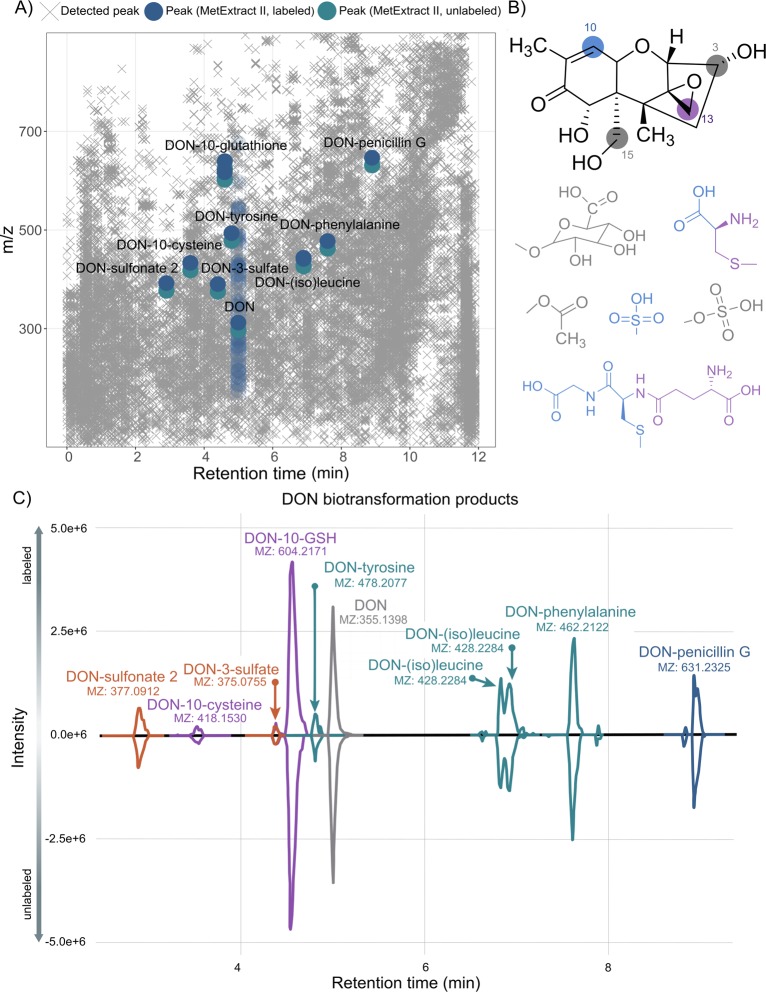
Figure 2.
Overview of data reduction by MetExtract, chemical structure and potential reaction sites of the food toxin DON, and detected biotransformation products. (A) All detected features in the analyzed samples are illustrated by gray ×’s (×). Features of metabolic products of deoxynivalenol detected by the MetExtract II algorithm are highlighted in blue, and features related to the parent molecule DON (eluting at 5 min) are shown in faded blue symbols (except for the [M + H]+ adduct) for better visibility. (B) Possible reaction sites for conjugation of DON. Positions C-3 and C-15 are targets for sulfate-, glucuronide-, and acetyl-conjugation. GSH as well as cysteine conjugates are formed at positions C-10 and C-13. DON-sulfonates are preferably formed by Michael addition at position C-10. (C) XIC chromatograms of annotated DON metabolites. The XICs of the native metabolite forms are shown with negative intensities, while the XICs of the 13C-labeled metabolite forms are shown with positive intensity values. The XICs were extracted from different raw data files, and for each metabolite the dominant adduct/in-source fragment is shown.