Fig. 3.
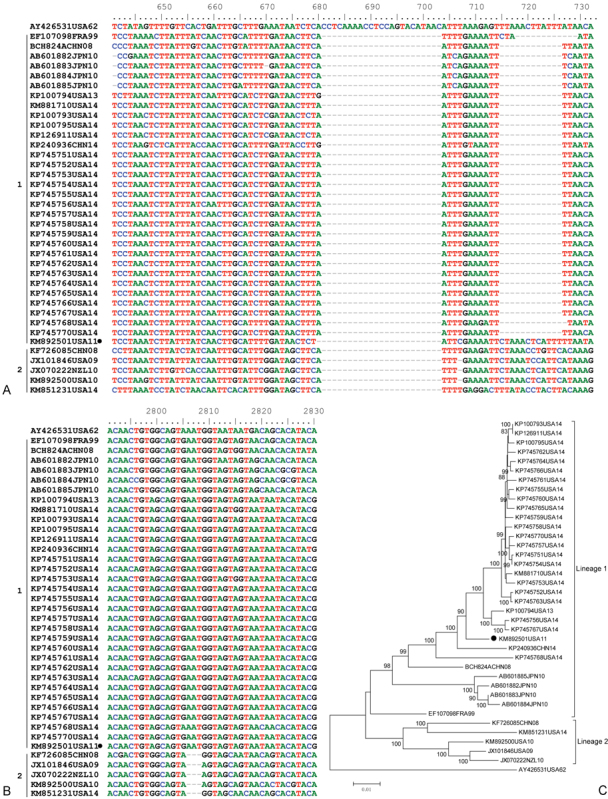
Nucletide alignment of all available complete EV-D68 genomes in GenBank. Nucletides are numbered relative to the start of the Fermon strain (AY426531) of EV-D68. The strains are indicated by Genbank accession numbers followed by state abbreviation and collection year. (A) Nucletide alignment of 5′UTR. Compared with the Fermon strain, all circulating strains have a 23 nt deletion at position 681–703. The strains of lineage 1 have an additional 11 nt deletion at position 705 to 726. (B) Nucletide alignment of VP1. Compared with the Fermon strain, strains of lineage 2 had 3 nt have a deletion at position 2,806 to 2,808. (C) Phylogenetic analysis of all available EV-D68 strains based on complete genomes. The trees, with 1,000 bootstrap replicates, were generated by using the neighbor-joining algorithm in MEGA 4.0. The strain has only one deletion as indicated by black circles.
