FIGURE 3. PARylation is at the intersection of primed and naïve pluripotent metabolomes.
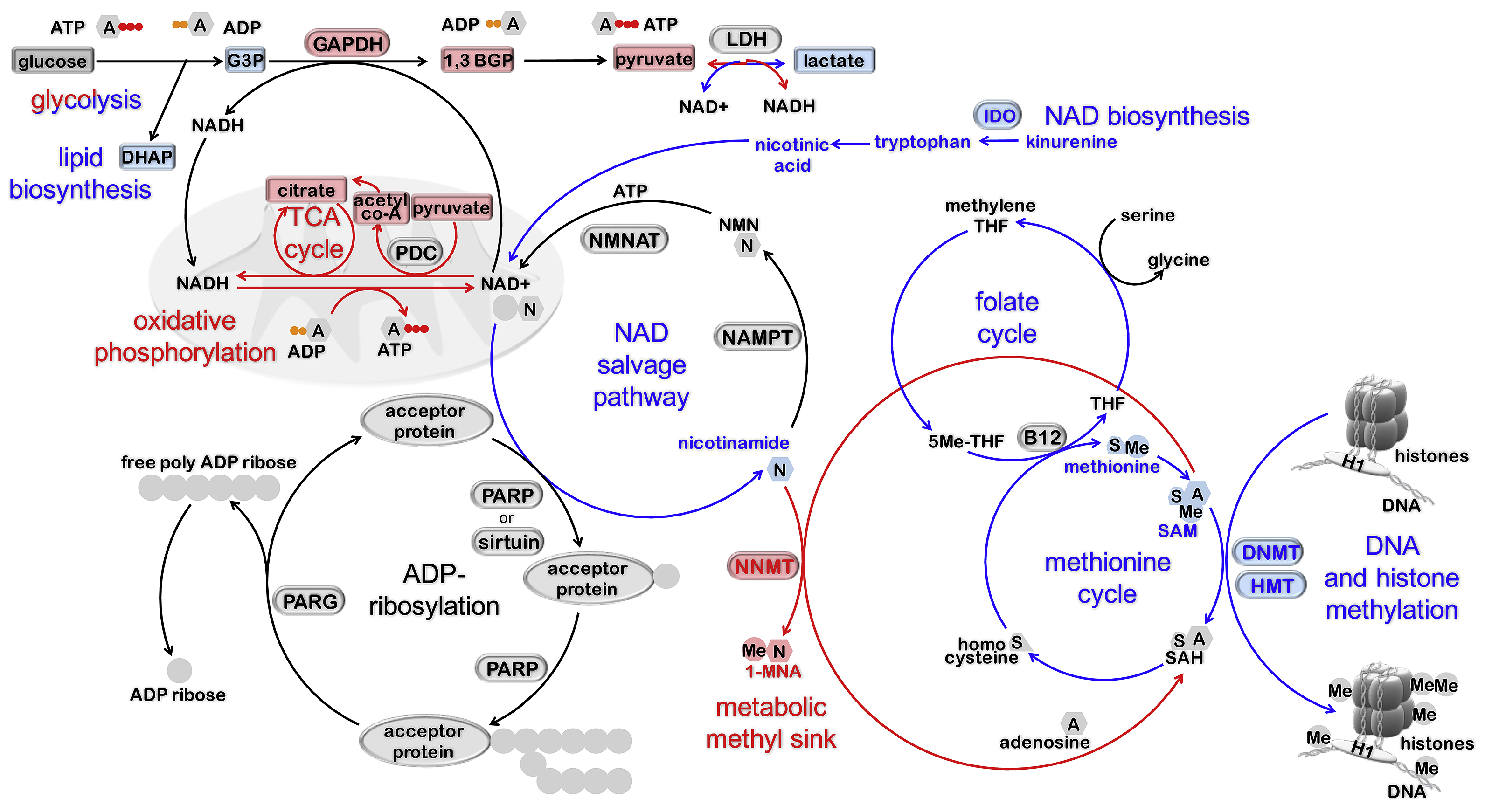
Distinct metabolomes have been characterized in primed and naïve states of human and mouse PSC [128, 129]. Metabolic activities, metabolites, and enzymes that are predominant in primed and naïve pluripotent states [128, 129] are indicated in blue and red, respectively. The substrate of ADP-ribosylation (i.e., NAD+) and its byproduct (i.e., nicotinamide) are integrated within PSC metabolic activities (i.e., oxidative phosphorylation, glycolysis, metabolic methyl-sink, one-carbon cycle). PARP-1, PARP-2, and tankyrases produce longer PAR polymers than all other PARP, and are responsible for most PARP-related NAD+ consumption. Thus, the PARP activity of these enzymes is presumptively the most affected by switching metabolism.
