Fig. 8. MiR-15a/16–1 translationally suppress mouse VEGFA, VEGFR2, FGF2 and FGFR1 in mouse brain microvascular endothelial cells.
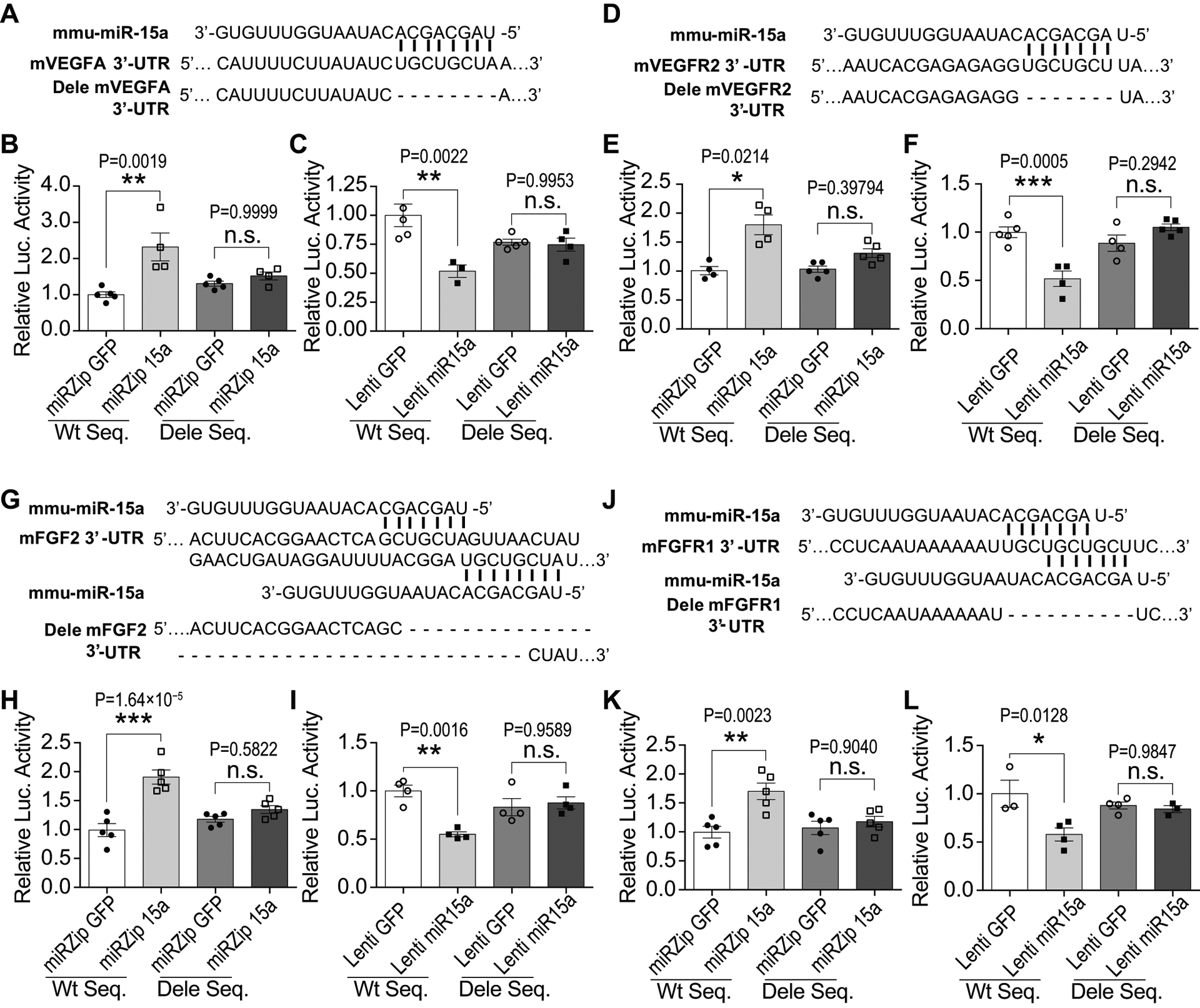
A,D,G,J, The partial sequence of mature mouse miR-15a (mmu-miR-15a) and its binding sites in the 3’-UTR region of the mouse VEGFA (A), VEGFR2 (D), FGF2 (G) and FGFR1 (J). MiR-15a reporter vectors containing CMV-driven expression of luciferase cDNA fused to mouse VEGFA (mVEGFA), mVEGFR2, mFGF2 and mFGFR1 wild-type 3’-UTR (WT Seq) or to miR-15a/16–1 binding site deleted 3’-UTR (the dash line nucleotides are deleted, Dele Seq) were constructed and transfected into bEnd3 cells. Meanwhile, bEnd3 cells were co-transduced with miRZip 15a or Lenti miR-15a for 72h prior to performing luciferase reporter activity assays. B,E,H,K, Quantitative data showed that loss-of-miR-15a function by miRZip 15a in bEnd3 cells statistically significantly increased luciferase activity of the reporter vector containing wild-type 3’-UTR sequence from mVEGFA (B), mVEGFR2 (E), mFGF2 (H) and mFGFR1 (K), in comparison with miRZip GFP. C,F,I,L, Gain-of-miR-15a function by Lenti miR-15a statistically significantly reduced luciferase activity of the reporter vector containing wild-type 3’-UTR sequence from mVEGFA (C), mVEGFR2 (F), mFGF2 (I) and mFGFR1 (L), in comparison with Lenti GFP. However, loss- or gain-of-miR-15a functions in bEnd3 cells had no effects on the luciferase activity of the reporter vector containing miR-15a binding site deleted 3’-UTR sequence (Dele Seq) from mouse VEGFA, VEGFR2, FGF2 or FGFR1. n = 3–5/group for all assays; *p < 0.05, **p < 0.01, ***p < 0.001 versus lentiviral GFP controls; n.s., no statistical significance; statistical analyses were performed by one-way ANOVA followed by Tukey’s multiple comparison tests for C, F, H, I, K, L; and statistical analyses were performed by Kruskal-Wallis test followed by Dunn’s multiple comparison tests for B and E.
