Abstract
The endoplasmic reticulum (ER) is the site of maturation for secretory and membrane proteins in eukaryotic cells. Unsuccessful folding attempts are eventually interrupted and most folding-defective polypeptides are dislocated across the ER membrane and degraded by cytosolic proteasomes in a complex series of events collectively defined as ER-associated degradation (ERAD). Uncontrolled ERAD activity might prematurely interrupt ongoing folding programs. At steady state, this is prevented by ERAD tuning, that is, the removal of select ERAD regulators from the ER and their degradation by proteasomes and by endo-lysosomal proteases. In Coronaviruses infected cells, the formation of LC3-I coated vesicles containing ERAD regulators cleared from the ER lumen is co-opted to anchor viral replication and transcription complexes to ER-derived membranes.
Introduction
Newly synthesized proteins emerge in the lumen of the endoplasmic reticulum (ER) from the Sec61 translocon. The translocon-associated protein oligosaccharyl transferase delays the folding of the nascent polypeptides to facilitate the transfer of oligosaccharides composed of three glucose, nine mannose and two N-acetylglucosamine residues (Figure 1 ) from a lipid donor in the ER membrane to the side chain of asparagines (N) in N–X–S/T or, more rarely, N–X–C sequons [1, 2, 3, 4]. Protein-bound oligosaccharides enhance solubility of unfolded nascent chains and facilitate protein maturation by recruiting ER-resident lectins and folding enzymes. Nevertheless, protein folding may fail and occasional or inherited amino acid mutations may substantially decrease the folding efficiency. Terminally misfolded polypeptides are deviated into an expanding number of specific ER-associated degradation (ERAD) pathways whose selection depends on biophysical features of the misfolded polypeptides such as the presence or the absence of N-glycans or of a transmembrane anchor. In the first part of the review, we will report on how processing of protein-bound oligosaccharides produces glycan structures that determine whether protein folding-attempts can be prolonged or must eventually be interrupted. We will also survey recent literature on the relationship between ERAD substrate topology and selection of ERAD pathways. In the second part, we will present the emerging concept of ERAD tuning. We propose that, at steady state, the efficient maturation of nascent cargo proteins might crucially depend on limitation of the ERAD capacity to avoid premature interruption of ongoing folding programs. This is obtained by segregating from the folding compartment select ERAD regulators, which in unstressed cells are characterized by much shorter half-life compared to conventional ER chaperones and folding enzymes.
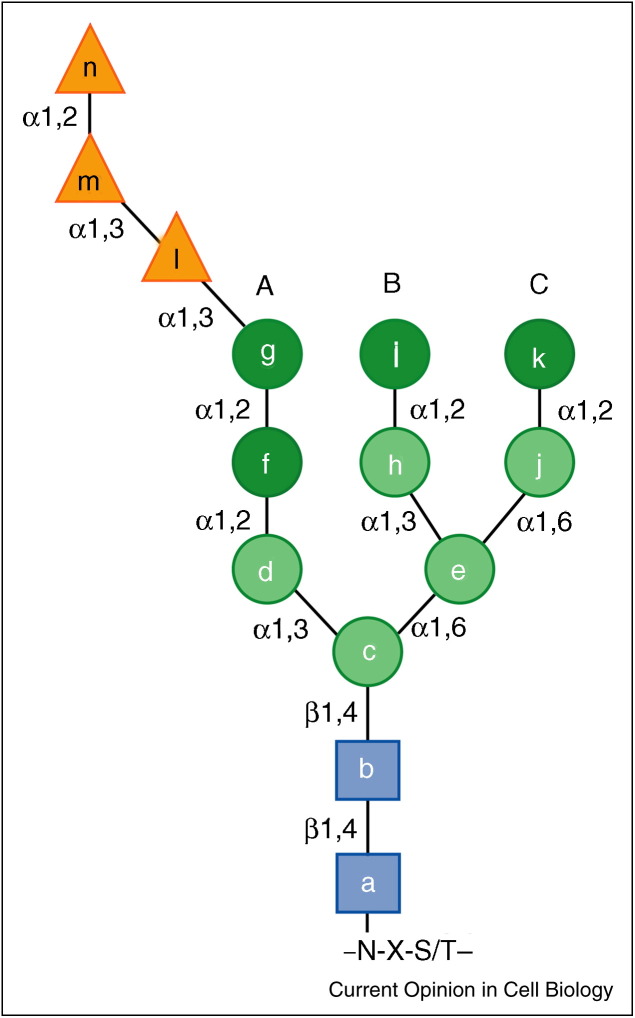
Figure 1.
The N-linked oligosaccharide structure. The core oligosaccharide is added onto side chains of asparagine residues in a specific sequon (N = asparagine, X = any amino acid but proline, S/T = serine or threonine). It is composed of three glucose (triangles), nine mannose (circles) and two N-acetylglucosamine (squares) residues. Removable α1,2-linked mannose residues are shown in dark green. Letters a-n are assigned to each saccharide and A–C define the oligosaccharide branch. The linkage between individual saccharides is shown.
Optimizing glycopolypeptide maturation: the role of ER-resident glucose processing and glucose binding proteins
Immediately after oligosaccharide addition onto nascent polypeptide chains, the two outermost glucose residues (n and m in Figure 1) are removed by the sequential intervention of the exoglucosidases I and II. Protein-linked mono-glucosylated oligosaccharides recruit a sophisticated folding device composed of the two lectin chaperones calnexin and calreticulin and an associated oxidoreductase, ERp57. ERp57 catalyzes a rate-determining step of glycoprotein folding, the formation of native intramolecular and intermolecular covalent bonds between cysteine residues (Figure 2 , step 1) [5, 6, 7]. Upon release from calnexin and calreticulin, the innermost glucose l is removed by the exoglucosidase II to prevent immediate re-association of the folding polypeptide with the ER-resident glucose-binding lectins. Most glycopolypeptides probably attain the native structure at this stage [8, 9]. For a subset of them, the intervention of specific cargo lectins facilitates export from the ER (Figure 2, step 2) [10]. Examples have been reported of polypeptides that undergo several cycles of release from/re-association with calnexin before attainment of the native structure [8, 11]. These cycles are driven by the folding sensor UDP-glucose:glycoprotein glucosyltransferase (UGT1) that specifically re-glucosylates the terminal mannose g of non-native polypeptides thus sending them back for another round of folding attempts in association with calnexin (D. Williams in this issue, Figure 2, step 3 [12, 13]).
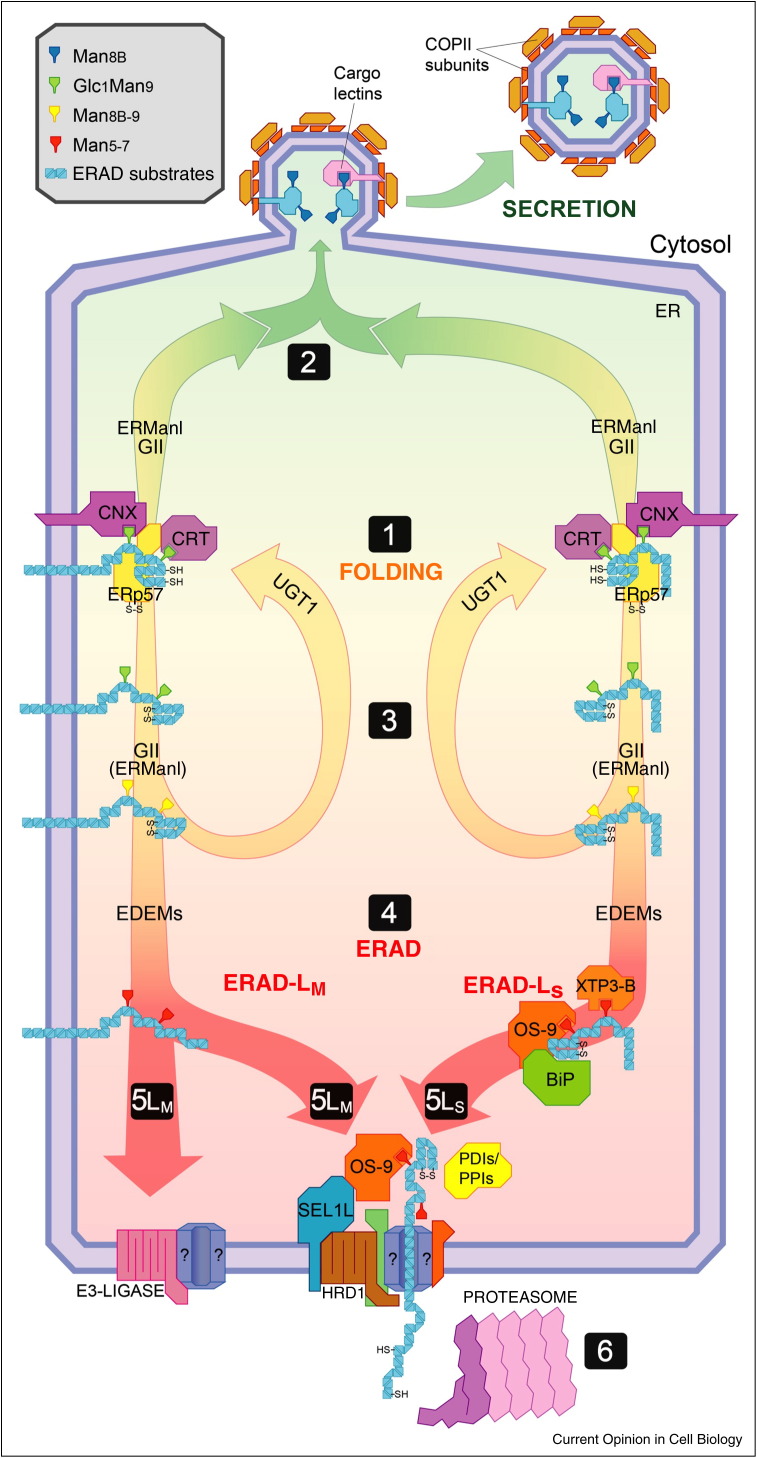
Figure 2.
Folding and ERAD pathways in the mammalian ER lumen. Newly synthesized glycopolypeptides associate with the lectin chaperones calnexin (CNX) and calreticulin (CRT). The oxidoreductase ERp57 catalyzes formation of native disulfide bonds (step 1). Upon release from CNX and CRT, the glucose l and the mannose i are removed by the exoglucosidase II (GII) and the ERManI, respectively. Native glycopolypeptides, in some cases under the assistance of specialized cargo lectins, are secreted in coat protein complex II (COPII)-coated vesicles and are transported at their final destination (step 2). Non-native glycopolypeptides are retained in the CNX chaperone system by the UGT1 that adds-back one glucose residue on the mannose residue g (step 3). Extensive de-mannosylation irreversibly extracts terminally misfolded polypeptides from the CNX cycle (step 4). Pathways directing ERAD substrates to dislocation sites at the ER membrane obligatorily rely on OS-9/XTP3-B, CyPB (for substrates containing peptidyl-prolyl bonds in the cis configuration), SEL1L and HRD1 only for ERAD-LS proteins (step 5LS). ERAD-LM substrates may engage multiple pathways (steps 5LM). Dislocation across the ER membrane occurs through an elusive proteinaceous channel (?). At the cytosolic face of the ER membrane ERAD substrates are poly-ubiquitylated, de-glycosylated and degraded by 26S-proteasomes (step 6).
Active interruption of unproductive folding attempts and generation of an ERAD signal: mechanisms conservation and possible divergences between Saccharomyces cerevisiae and other Eukarya
Even for correct gene products, a fraction ranging from 30% [14] to much less [15] never attains the native structure. Folding-defective polypeptides are cleared from the ER lumen and most of them are degraded by cytosolic proteasomes. The series of events leading to disposal of terminally misfolded glyco-polypeptides from the ER lumen have initially been characterized in the budding yeast S. cerevisiae [16, 17]. The yeast ERManI first removes the mannose residue i (Figure 1) from protein-bound oligosaccharides. This is not sufficient to tag polypeptides for disposal because it also occurs for native polypeptides that will be selected for secretion (Figure 2, step 2) [18]. However, it is a pre-requisite for the intervention of yeast EDEM, which removes the mannose residue k (Figure 1) from oligosaccharides displayed on misfolded conformers [19, 20•, 21•]. This cleavage generates a signal that recruits Yos9p, an ERAD lectin containing a mannose 6-phosphate receptor homology domain that binds Man7 oligosaccharides exposing the terminal α1,6-bonded mannose j [20•, 21•, 22]. Yos9p releases misfolded polypeptides with luminal or transmembrane folding defects (ERAD-L and ERAD-M substrates) to retro-translocation complexes built around the membrane-embedded E3 ubiquitin ligase Hrd1p. Misfolded polypeptides with cytosolic folding defects (ERAD-C substrates) engage retro-translocation complexes containing the E3 ubiquitin ligase Doa10p. These complexes facilitate transport of misfolded polypeptides across the ER membrane and their proteasomal degradation [23, 24, 25, 26, 27, 28, 29, 30].
The folding sensor UGT1, which is absent in S. cerevisiae, determines the quality control pathways operating in most other Eukarya. UGT1 could indefinitely delay ERAD onset by continual re-glucosylation of the oligosaccharide branch A (Figure 1). This would prevent release of folding-defective polypeptides from the calnexin chaperone system (Figure 2, step 3). Unlike S. cerevisiae, more extensive de-mannosylation of folding-defective polypeptides with removal of all α1,2-linked mannose residues has been reported for mammalian cells (Figure 1, dark green) [31]. In particular, the removal of mannose residues g and f is documented [2, 32, 33] and requires the intervention of several members of the glycosyl hydrolase family 47 comprising the ER-mannosidase I (ERManI), EDEM1, EDEM2 and EDEM3 and, possibly, also the intervention of endo-mannosidases (Figure 2, step 4) [34]. All in all, extensive de-mannosylation of ERAD candidates in the mammalian ER reduces the efficiency of UGT1 re-glucosylation, eventually removes the glucose acceptor-site (mannose g) and generates a signal that may elicit intervention of ERAD lectins, which direct terminal misfolded polypeptides to specialized dislocation sites at the ER membrane (steps 5LM and 5LS).
Substrate-specific ERAD pathways: ERAD-LS and ERAD-LM substrates
Cumulating data acquired by monitoring the fate of an increasing number of model ERAD substrates highlight the complexity of the quality control mechanisms operating to ensure most efficient recognition and clearance of aberrant polypeptides from the mammalian ER [35]. The aggregation proneness of the misfolded polypeptides determines whether ERAD or autophagic pathways are activated for disposal [36]. For ERAD substrates, the presence of protein-bound oligosaccharides determines the intervention of sugar processing and sugar binding ER proteins, while their absence results in activation of quality control pathways that are much less understood [2].
Recent systematic studies of glycopolypeptides characterized by structural defects in their luminal portion and modified to add (ERAD-LM substrates) or to delete (ERAD-LS substrates) a membrane anchor revealed unsuspected mechanistic variations in the ERAD pathways for glycoproteins (Figure 2, step 5LM versus step 5LS) [37•]. While confirming the requirement of extensive de-mannosylation to interrupt unproductive folding attempts and the intervention of cytosolic proteasomes for degradation, these studies showed that only the ERAD-LS substrates strictly require the intervention of the ERAD lectins OS-9 and/or XTP3-B, of the membrane adaptor SEL1L and of the E3 ubiquitin ligase HRD1 [37•]. Moreover, disposal of ERAD-LS substrates containing peptidyl-prolyl bonds in the cis configuration (but not of their ERAD-LM counterparts) is inhibited by Cyclosporine A and requires the enzymatic intervention of the luminal peptidyl-prolyl isomerase cyclophilin B (CyPB) [38]. The cis to trans isomerization of peptidyl-prolyl bonds may facilitate dislocation of ERAD candidates across the ER membrane by promoting their detachment from luminal chaperones. Alternatively, like the reduction of disulfide bonds [39], it could facilitate the dislocation through the elusive, membrane-embedded retro-translocation channel (? in Figure 2) by eliminating turns in the misfolded polypeptide secondary structures ([38] and Figure 2, step 5LS). Requirement for OS-9/XTP3-B, CyPB, SEL1L and HRD1 interventions is less stringent for efficient disposal of the same polypeptides when they are anchored at the membrane (ERAD-LM substrates, Figure 2, step 5LM [37•, 38]). It can be envisioned that membrane-bound misfolded polypeptides may eventually access dislocation sites by lateral diffusion in the ER membrane, while for soluble polypeptides an active transport from the ER lumen to the membrane-embedded dislocons is crucial for the efficient clearance from the ER [38]. In this context, it is important to mention that unlike other E3 ubiquitin ligases, the HRD1 is characterized by the association of adaptors that recruit luminal factors such as the ERAD lectins OS-9 and XTP3-B that may act as shuttles to transfer soluble ERAD substrates from the folding machinery to the dislocation sites at the ER membrane. The mechanisms that regulate the handoff of ERAD substrates are unclear and data showing that EDEM1 [40•] and OS-9 [41•] use their lectin sites to form complexes with components of the dislocation machinery (i.e. SEL1L) rather than with misfolded proteins raise new questions on the actual role of protein-bound oligosaccharides in ERAD.
A competition for non-native proteins: loss-of-function disorders
How folding intermediates to be preserved are distinguished from terminally misfolded conformers to be degraded remains a central question in the field. Several observations indicate that conformational maturation and selection for disposal are in kinetic competition in the ER lumen [42]. For example, many loss-of-function genetic diseases are caused by mutations that do not affect the function of the polypeptide, but delay the folding process such that immature conformers are degraded before attainment of the native structure. In such cases, chemical or pharmacological chaperones that promote maturation before the onset of polypeptide disposal can rescue the disorder [42, 43]. Interestingly, the enhanced expression of ERAD regulators such as ERManI [44], EDEM proteins [45] or E3 ubiquitin ligases [46] may result in the premature destruction of folding intermediates. On the other hand the reduction of the intralumenal level of ERAD regulators or their pharmacologic inactivation offers additional time to cargo proteins characterized by slow maturation to attain the native structure. This enhances folding and secretion efficiency and may alleviate disease phenotypes [46, 47, 48, 49, 50, 51, 52].
Life and death of ERAD regulators: ERAD tuning
In the ER lumen, non-native folding intermediates must be protected from unwanted recognition by ERAD regulators that could prematurely interrupt ongoing folding programs. One model claims that this is obtained upon compartmentalization of the disposal machinery in sub-regions of the ER characterized by high ERManI content, the ER quality control compartment (ERQC [31]). Terminally misfolded proteins would be transported at the ERQC to be subject to the extensive de-mannosylation that tags them for disposal [2, 31]. However, emerging evidences show that the selective degradation of ERAD regulators in a series of events collectively termed ERAD tuning may contribute to the reduction of the ERAD capacity at levels that do not interfere with maturation of newly synthesized cargo proteins at steady state [45]. Our model is based on data showing that several folding factors (e.g. calnexin, calreticulin, BiP, PDI, ERp57, ERp72 and GRP94) are long-living proteins [45, 53], while many ERAD regulators (e.g. ERManI [54, 55], EDEM1 [45, 56, 57•], OS-9 [57•], XTP3-B [58], HERP [53, 59] and SEL1L [60]) are rapidly removed from the ER in unstressed cells. HERP that contains an ubiquitin-like domain and SEL1L are degraded by the proteasome [53, 59, 60]; the ERManI [54, 55], EDEM1 [45, 56, 57•] and OS-9 [57•] by endosomal/lysosomal enzymes (Figure 3 ).
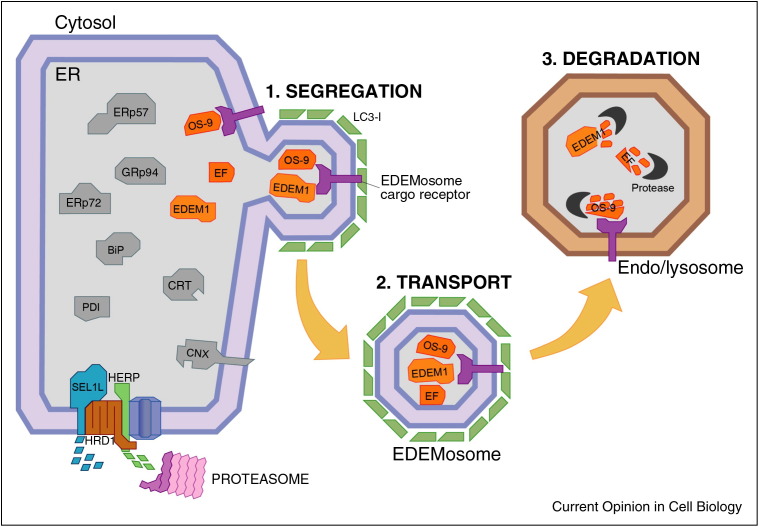
Figure 3.
ERAD tuning. Many ERAD regulators are short-living proteins at steady state. Some of them are degraded with the intervention of cytosolic proteasomes (e.g. SEL1L and HERP). The selective removal of EDEM1 and OS-9 from the ER can be subdivided in three steps. (1) Association with an elusive receptor allows segregation of EDEM1, OS-9 and possibly other ERAD factors (EF) from conventional, long living ER-resident chaperones (in grey); (2) the ERAD regulators exit the ER in small, LC3-I-coated vesicles, the EDEMosomes; (3) EDEMosomes deliver their content to endosomal/lysosomal compartments for disposal.
Subcellular fractionation [45] and electron microscopy studies [61] revealed that at steady state about 80% of the endogenous EDEM1 localizes in small ER-derived vesicles, the EDEMosomes. Originally, these vesicles were thought to be involved in the removal of misfolded cargo proteins from the ER lumen [61]. However, our studies showing that EDEM1 and OS-9 do accumulate in these vesicles when cells are exposed to lysosomotropic drugs or are infected with Coronaviruses (see below) revealed that EDEMosomes are rather involved in the clearance of short-living ERAD regulators from the ER lumen [45, 57•, 62]. The rapid turnover of EDEM1 and OS-9 relies on firstly, their segregation from long-living ER chaperones; secondly, their export from the ER in small vesicles; and thirdly, their degradation in endosomal/lysosomal compartments [45, 57•] (Figure 3). The non-covalent association of the ubiquitin-like protein LC3 at the cytosolic face of the membrane distinguishes EDEMosomes from autophagosomes [45], which display LC3 covalently bound to membrane lipids [63] and from secretory vesicles, which display a COPII-coat [10]. Although some controversy still exists [56] and some component of the autophagy machinery participates in the process, ERAD tuning is clearly distinct from macroautophagy [45, 57•]. With such regulatory mechanisms operating in the ER, adaptation of ERAD activity to transient accumulation of aberrant polypeptides in the ER lumen might not necessarily await the activation of the transcriptional unfolded protein response (UPR) programs [64]. Rather, it is conceivable that ERAD activity can rapidly be modulated ‘on demand’ when association with misfolded conformers retains ERAD regulators in the ER lumen thus preventing their rapid segregation from the compartment that occurs at steady state.
Conclusions and perspectives
The capacity to intervene in protein biogenesis by improving the rate and/or the efficiency of protein folding and by modulating the degradation of non-native polypeptides has important clinical and industrial implications.
The rapid degradation of select ERAD regulators (ERAD tuning) may contribute in determining the basal level of ERAD activity. At steady state, basal ERAD must insure disposal of by-products of protein biogenesis that would otherwise progressively accumulate, without interfering with ongoing folding programs. Physiologic or pathologic variations in the level of misfolded polypeptides may require enhancement of the ERAD capacity. This can be obtained upon the well-studied induction of an UPR transcriptional program, which may take several hours and may eventually lead to cell death if recovery is impossible [65]. Alternatively, a more rapid and readily reversible ERAD enhancement could rely on the shutdown of ERAD tuning occurring when ERAD regulators that are normally rapidly degraded do actually remain in the ER upon association with accumulating misfolded conformers that need assistance.
The activation of autophagy inhibits disposal of EDEM1 and of OS-9 [45, 57•] possibly because LC3-I, which is involved in their vesicular export from the ER, is converted in autophagosomal-membrane-bound LC3-II [63]. If inhibition of this branch of ERAD tuning (the other branch relies on degradation of ERAD regulators by cytosolic proteasomes, Figure 3) is sufficient to enhance the overall ERAD activity, one could envision interesting medical implications. The cross talk between autophagy and ERAD tuning may for example contribute to delay the progression of diseases such as serpinopathies where the accumulation of misfolded polypeptides does not induce an UPR [66, 67]. In such cases, the enhancement of the ERAD activity to contrast the accumulation of misfolded conformers could solely depend on the inhibition of ERAD tuning.
Since the machineries regulating folding and degradation of proteins entering the secretory pathway are up-regulated in many types of tumors and are hijacked in many ways by pathogens of bacterial and viral origin, the detailed mechanistic characterization of the events described in this review will hopefully offer new targets for therapeutic intervention. In the context of this review, the characterization of the vesicular pathway exiting the ER to reduce the luminal content of select ERAD factors seems important since at least one class of pathogens, the Coronaviruses, has been identified that co-opts the ERAD tuning machinery. In fact, in infected cells, the EDEMosomes or a modified version thereof containing EDEM1 and OS-9, host the viral replication and transcription complexes. Consistently, viral infection interferes with ERAD tuning and results in accumulation of EDEMosomal cargo in the viral replicosomes [45, 57•, 62].
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgments
We would like to thank L. Ruddock for insightful comments on the manuscript. S. Bianchi is acknowledged for Figure 2. M.M. is supported by grants from the Foundation for Research on Neurodegenerative Diseases, the Fondazione San Salvatore, the Swiss National Center of Competence in Research on Neural Plasticity and Repair, the Swiss National Science Foundation and ONELIFE Advisors SA.
References
- 1.Helenius A., Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 2.Aebi M., Bernasconi R., Clerc S., Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Wilson C.M., High S. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J Cell Sci. 2007;120:648–657. doi: 10.1242/jcs.000729. [DOI] [PubMed] [Google Scholar]
- 4.Schulz B.L., Stirnimann C.U., Grimshaw J.P., Brozzo M.S., Fritsch F., Mohorko E., Capitani G., Glockshuber R., Grutter M.G., Aebi M. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci U S A. 2009;106:11061–11066. doi: 10.1073/pnas.0812515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zapun A., Darby N.J., Tessier D.C., Michalak M., Bergeron J.J., Thomas D.Y. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 6.Oliver J.D., van der Wal F.J., Bulleid N.J., High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 7.Soldà T., Garbi N., Hammerling G.J., Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- 8.Soldà T., Galli C., Kaufman R.J., Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–249. doi: 10.1016/j.molcel.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 9.Fanchiotti S., Fernandez F., D’Alessio C., Parodi A.J. The UDP-Glc:glycoprotein glucosyltransferase is essential for Schizosaccharomyces pombe viability under conditions of extreme endoplasmic reticulum stress. J Cell Biol. 1998;143:625–635. doi: 10.1083/jcb.143.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dancourt J., Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 11.Pearse B.R., Tamura T., Sunryd J.C., Grabowski G.A., Kaufman R.J., Hebert D.N. The role of UDP-Glc:glycoprotein glucosyltransferase 1 in the maturation of an obligate substrate prosaposin. J Cell Biol. 2010;189:829–841. doi: 10.1083/jcb.200912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Totani K., Ihara Y., Tsujimoto T., Matsuo I., Ito Y. The recognition motif of the glycoprotein-folding sensor enzyme UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 2009;48:2933–2940. doi: 10.1021/bi8020586. [DOI] [PubMed] [Google Scholar]
- 13.Caramelo J.J., Parodi A.J. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 15.Vabulas R.M., Hartl F.U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 16.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stolz A., Wolf D.H. Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochim Biophys Acta. 2010;1803:694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Moremen K.W. In: Ernst B., Hart G., Sinay P., editors. vol 2. John Wiley and Sons, Inc.; New York: 2000. (Alpha-Mannosidases in Asparagine-Linked Oligosaccharide Processing and Catabolism). [Google Scholar]
- 19.Xie W., Kanehara K., Sayeed A., Ng D.T. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell. 2009;20:3317–3329. doi: 10.1091/mbc.E09-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Quan E.M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J.S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Reference [21•].
- 21•.Clerc S., Hirsch C., Oggier D.M., Deprez P., Jakob C., Sommer T., Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that yeast EDEM is an active mannosidase that generates an oligosaccharide structure recognized by Yos9p as a crucial signal that directs misfolded polypeptides for degradation.
- 22.Mikami K., Yamaguchi D., Tateno H., Hu D., Qin S.Y., Kawasaki N., Yamada M., Matsumoto N., Hirabayashi J., Ito Y. The sugar-binding ability of human OS-9 and its involvement in ER-associated degradation. Glycobiology. 2010;20:310–321. doi: 10.1093/glycob/cwp175. [DOI] [PubMed] [Google Scholar]
- 23.Kanehara K., Xie W., Ng D.T. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol. 2010;188:707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer M., Forte G.M., Stirling C.J. Sec61p is required for ERAD-L: genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J Biol Chem. 2008;283:33883–33888. doi: 10.1074/jbc.M803054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauss R., Jarosch E., Sommer T., Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 26.Denic V., Quan E.M., Weissman J.S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho P., Goder V., Rapoport T.A. Distinct ubiquitin–ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 28.Vashist S., Ng D.T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huyer G., Longsworth G.L., Mason D.L., Mallampalli M.P., McCaffery J.M., Wright R.L., Michaelis S. A striking quality control subcompartment in Saccharomyces cerevisiae: the endoplasmic reticulum-associated compartment. Mol Biol Cell. 2004;15:908–921. doi: 10.1091/mbc.E03-07-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taxis C., Hitt R., Park S.H., Deak P.M., Kostova Z., Wolf D.H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- 31.Lederkremer G.Z. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Olivari S., Cali T., Salo K.E., Paganetti P., Ruddock L.W., Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- 33.Ermonval M., Kitzmuller C., Mir A.M., Cacan R., Ivessa N.E. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 2001;11:565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- 34.Olivari S., Molinari M. Glycoprotein folding and the role of EDEM1. EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 2007;581:3658–3664. doi: 10.1016/j.febslet.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 35.Brodsky J.L., Wojcikiewicz R.J. Substrate-specific mediators of ER associated degradation (ERAD) Curr Opin Cell Biol. 2009;21:516–521. doi: 10.1016/j.ceb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlmutter D.H. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2006;2:258–263. doi: 10.4161/auto.2882. [DOI] [PubMed] [Google Scholar]
- 37•.Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study and reference [38] show that differences in biophysical properties such as the topology may affect the selection of ERAD factors and pathways activated for efficient removal of misfolded proteins from the mammalian ER.
- 38.Bernasconi R., Soldà T., Galli C., Pertel T., Luban J., Molinari M. Cyclosporine A-sensitive, cyclophillin B-dependent endoplasmic reticulum-associated protein degradation. PLoS ONE. 2010;5:e13008. doi: 10.1371/journal.pone.0013008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellgaard L., Ruddock L.W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Cormier J.H., Tamura T., Sunryd J.C., Hebert D.N. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Reference [41•].
- 41•.Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1/SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]; By showing that the lectin sites of EDEM1 and of OS-9 do associate with SEL1L oligosaccharides, this study opens new possibilities on roles of protein-bound oligosaccharides in ERAD regulation.
- 42.Hutt D., Balch W.E. Cell biology. The proteome in balance. Science. 2010;329:766–767. doi: 10.1126/science.1194160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatahet F., Ruddock L.W. Modulating proteostasis: peptidomimetic inhibitors and activators of protein folding. Curr Pharm Des. 2009;15:2488–2507. doi: 10.2174/138161209788682343. [DOI] [PubMed] [Google Scholar]
- 44.Wu Y., Swulius M.T., Moremen K.W., Sifers R.N. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc Natl Acad Sci U S A. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cali T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun. 2008;371:405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 46.Younger J.M., Chen L., Ren H.Y., Rosser M.F., Turnbull E.L., Fan C.Y., Patterson C., Cyr D.M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 47.Vanoni O., Paganetti P., Molinari M. Consequences of individual N-glycan deletions and of proteasomal inhibition on secretion of active BACE. Mol Biol Cell. 2008;19:4086–4098. doi: 10.1091/mbc.E08-05-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vij N., Fang S., Zeitlin P.L. Selective inhibition of endoplasmic reticulum-associated degradation rescues DeltaF508-cystic fibrosis transmembrane regulator and suppresses interleukin-8 levels: therapeutic implications. J Biol Chem. 2006;281:17369–17378. doi: 10.1074/jbc.M600509200. [DOI] [PubMed] [Google Scholar]
- 49.Farinha C.M., Amaral M.D. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol Cell Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonuccelli G., Sotgia F., Schubert W., Park D.S., Frank P.G., Woodman S.E., Insabato L., Cammer M., Minetti C., Lisanti M.P. Proteasome inhibitor (MG-132) treatment of mdx mice rescues the expression and membrane localization of dystrophin and dystrophin-associated proteins. Am J Pathol. 2003;163:1663–1675. doi: 10.1016/S0002-9440(10)63523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grove D.E., Rosser M.F., Ren H.Y., Naren A.P., Cyr D.M. Mechanisms for rescue of correctable folding defects in CFTRDelta F508. Mol Biol Cell. 2009;20:4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcus N.Y., Perlmutter D.H. Glucosidase and mannosidase inhibitors mediate increased secretion of mutant alpha1 antitrypsin Z. J Biol Chem. 2000;275:1987–1992. doi: 10.1074/jbc.275.3.1987. [DOI] [PubMed] [Google Scholar]
- 53.Hori O., Ichinoda F., Yamaguchi A., Tamatani T., Taniguchi M., Koyama Y., Katayama T., Tohyama M., Stern D.M., Ozawa K. Role of Herp in the endoplasmic reticulum stress response. Genes Cells. 2004;9:457–469. doi: 10.1111/j.1356-9597.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- 54.Termine D.J., Moremen K.W., Sifers R.N. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J Cell Sci. 2009;122:976–984. doi: 10.1242/jcs.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y., Termine D.J., Swulius M.T., Moremen K.W., Sifers R.N. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Fourn V., Gaplovska-Kysela K., Guhl B., Santimaria R., Zuber C., Roth J. Basal autophagy is involved in the degradation of the ERAD component EDEM1. Cell Mol Life Sci. 2009;66:1434–1445. doi: 10.1007/s00018-009-9038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Reggiori F., Monastyrska I., Verheije M.H., Cali T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that the ER-derived vesicles involved in replication of Coranaviruses do not contain conventional ER chaperones but EDEM1 and OS-9 whose rapid intracellular turnover (ERAD tuning as defined in [45]) is blocked upon viral infection.
- 58.Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miura H., Hashida K., Sudo H., Awa Y., Takarada-Iemata M., Kokame K., Takahashi T., Matsumoto M., Kitao Y., Hori O. Deletion of Herp facilitates degradation of cytosolic proteins. Genes Cells. 2010;15:843–853. doi: 10.1111/j.1365-2443.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- 60.Mueller B., Lilley B.N., Ploegh H.L. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber C., Cormier J.H., Guhl B., Santimaria R., Hebert D.N., Roth J. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc Natl Acad Sci U S A. 2007;104:4407–4412. doi: 10.1073/pnas.0700154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Haan C.A., Molinari M., Reggiori F. Autophagy-independent LC3 function in vesicular traffic. Autophagy. 2010;6:994–996. doi: 10.4161/auto.6.7.13309. [DOI] [PubMed] [Google Scholar]
- 63.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kincaid M.M., Cooper A.A. ERADicate ER stress or die trying. Antioxid Redox Signal. 2007;9:2373–2387. doi: 10.1089/ars.2007.1817. [DOI] [PubMed] [Google Scholar]
- 65.Rutkowski D.T., Hegde R.S. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hidvegi T., Schmidt B.Z., Hale P., Perlmutter D.H. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J Biol Chem. 2005;280:39002–39015. doi: 10.1074/jbc.M508652200. [DOI] [PubMed] [Google Scholar]
- 67.Graham K.S., Le A., Sifers R.N. Accumulation of the insoluble PiZ variant of human alpha 1-antitrypsin within the hepatic endoplasmic reticulum does not elevate the steady-state level of grp78/BiP. J Biol Chem. 1990;265:20463–20468. [PubMed] [Google Scholar]