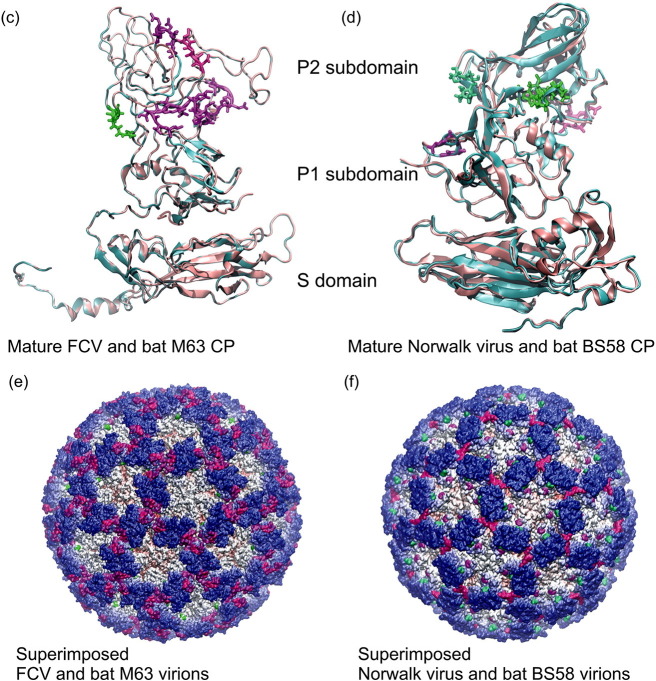
Fig. 1.
Structure comparison of mature calicivirus (ORF2, VP1 segment) CP proteins and virions. Structure based amino acid sequence alignments of the newly reported bat calicivirus capsid proteins and the template FCV and Norwalk calicivirus VP1 proteins (a) and (b). The background of the sequence alignments reflects the homology levels of the two–two related capsid protein sequences: identical amino acids are red, similar aas. are light orange while different aas. are light pink. The main structural differences are indicated by shades of magenta and green color codes on the sequence alignment and on the superimposed CP structures. The template calicivirus VP1 protein structures are illustrated in cyan cartoon representation, while the new bat calicivirus VP1 model structure are pink (c) and (d). Molecular surface representation of the superimposed template and the newly described bat calicivirus virions (e) and (f). The molecular surface is colored by radial extension of the amino acids from the virion center. Dark blue represents the most protruding CP parts. The structural differences were colored in the same way as for the capsid monomers. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)