Fig. 1.
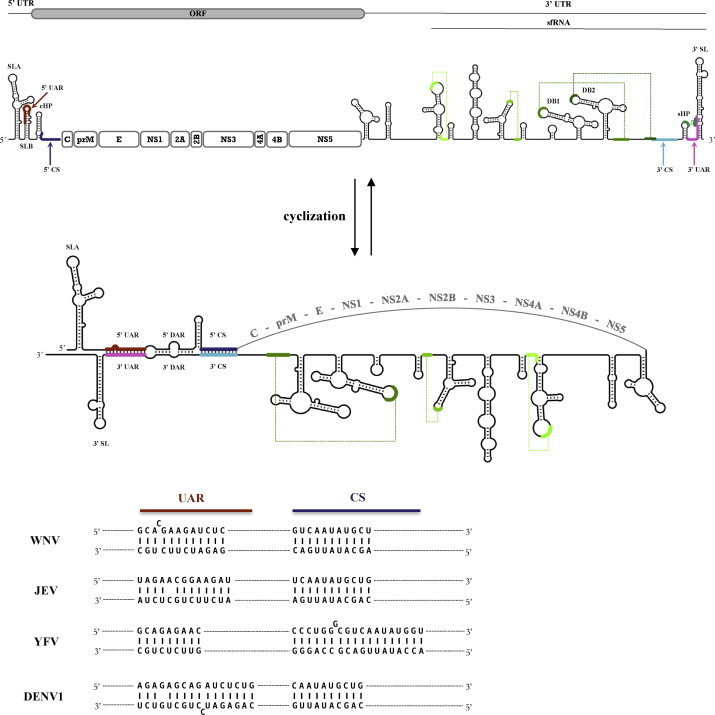
Schematic representation of the linear and circular forms of WNV genomic RNA. Flavivirus infected cells accumulate two major RNA species, the viral genomic RNA and the small sfRNA (subgenomic flavivirus RNA). RNA secondary structures were predicted by Mfold (Zuker, 2003), using constraints on experimentally characterized RNA structures (see references in text). The conserved RNA structures and sequences include: SLA—stem-loop A; UAR—upstream AUG region; DAR—downstream AUG region; CS—conserved sequence; cHP—capsid region hairpin; sHP—small hairpin; DB1 and DB2—dumbbell structures; 3′ SL—3′ stem-loop. 5′ and 3′ UAR sequences are highlighted in dark red and magenta, respectively. 5′ and 3′ CS elements are highlighted in dark and light blue. Conserved pseudoknot structures are illustrated in green. The core protein-coding sequence partly overlaps with RNA secondary structures in the 5′ region of the genome. Note that the lower stem of 3′ SL is opened in the circular RNA form, facilitating binding of the viral replication complex. Cyclization sequences of representative mosquito-borne flaviviruses are shown below the circular RNA structure (GenBank accession for West Nile virus—AF260968; Japanese encephalitis virus—NC_001437.1; yellow fever virus—NC_002031.1; dengue virus type 1—NC_001477). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
