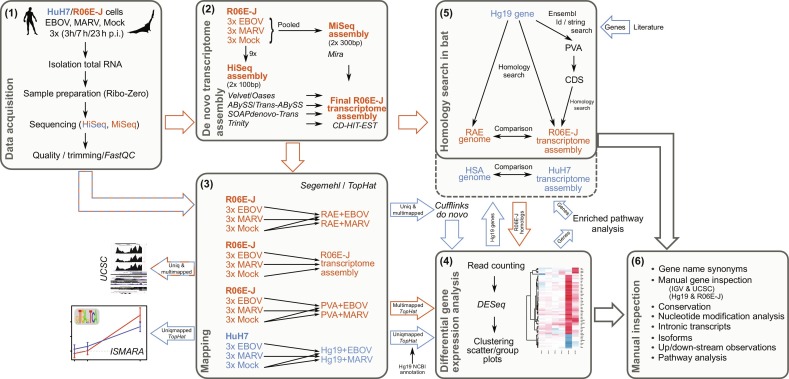
Fig. 5.
Host–virus RNA-Seq methods pipeline for the detection of differential expressed genes (see text for details).
Modified from Hölzer , M., Krähling, V., Amman, F., Barth, E., Bernhart, S.H., Carmelo, V.A.O., Collatz, M., Doose, G., Eggenhofer, F., Ewald, J., Fallmann, J., Feldhahn, L.M., Fricke, M., Gebauer, J., Gruber, A.J., Hufsky, F., Indrischek, H., Kanton, S., Linde, J., Mostajo, N., Ochsenreiter, R., Riege, K., Rivarola- Duarte, L., Sahyoun, A.H., Saunders, S.J., Seemann, S.E., Tanzer, A., Vogel, B., Wehner, S., Wolfinger, M.T., Backofen, R., Gorodkin, J., Grosse, I., Hofacker, I., Hoffmann, S., Kaleta, C., Stadler, P.F., Becker, S., Marz, M., 2016. Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells. Sci. Rep. 6, 34589. doi:10.1038/srep34589.