Abstract
Mitochondria are involved in multiple cellular functions, in addition to their core role in energy metabolism. Mitochondria localized in different cellular locations may have different morphology, Ca2+ handling and biochemical properties and may interact differently with other intracellular structures, causing functional specificity. However, most prior studies have utilized isolated mitochondria, removed from their intracellular environment. Mitochondria in cardiac ventricular myocytes are highly organized, with a majority squeezed between the myofilaments in longitudinal chains (intrafibrillar mitochondria, IFM). There is another population of perinuclear mitochondria (PNM) around and between the two nuclei typical in myocytes. Here, we take advantage of live myocyte imaging to test for quantitative morphological and functional differences between IFM and PNM with respect to calcium fluxes, membrane potential, sensitivity to oxidative stress, shape and dynamics. Our findings show higher mitochondrial Ca2+ uptake and oxidative stress sensitivity for IFM vs. PNM, which may relate to higher local energy demand supporting the contractile machinery. In contrast to IFM which are remarkably static, PNM are relatively mobile, appear to participate readily in fission/fusion dynamics and appear to play a central role in mitochondrial genesis and turnover. We conclude that while IFM may be physiologically tuned to support local myofilament energy demands, PNM may be more critical in mitochondrial turnover and regulation of nuclear function and import/export. Thus, important functional differences are present in intrafibrillar vs. perinuclear mitochondrial subpopulations.
Keywords: mitochondrial Ca, mitochondrial dynamic, mitochondrial heterogeneity
Introduction
Mitochondria localized in different regions of a cell, interact with local intracellular structures and may display distinct morphological and biochemical properties. These mitochondrial subsets may also exhibit different responses to substrates, inhibitors and may vary in their energetics, sensitivity to stimulation, resistance to apoptosis, and other characteristics. In cardiac mycoytes, mitochondria are spatially separated into different populations. One just beneath the surface sarcolemma (subsarcolemmal mitochondria, SSM), the major population which are between the myofibrils (interfibrillar mitochondria, IFM), and a third population that is perinuclear (PNM). The IFM are organized in a lattice of parallel rows surrounding and surrounded by the contractile myofilament and these IFM may be restricted in their positions. This type of organization may provide bioenergetic support for contraction and mitochondrial interactions with the cytoskeleton [1, 2] and sarcoplasmic reticulum (SR) [3–5]. The heart performs energetically demanding work (contraction, rapid cycles of ion transport) that require large amounts of ATP every second and that work can dynamically regulate gene expression and protein synthesis to adapt to different chronic workloads.
In adult cardiac myocytes, the precise matching of energy supply with demand are critical in maintaining cardiac function at different workloads and substrate supply. Heterogeneity of mitochondrial energetics can cause complex dynamic behavior to deal with these changes in cellular workload or environmental conditions [6]. This includes local metabolic transients, cell-wide coordinated redox transitions, and propagated metabolic waves within myocytes [7]. There is growing evidence for functional heterogeneity of mitochondria with respect to their biochemical, respiratory or enzymatic activities, Ca2+ handling and membrane potential. However most prior characterizations have relied on isolated mitochondria that are separated from their normal in-situ environment [8–12], which may limit conclusions. Confocal imaging allows spatial analysis of different mitochondrial populations in live myocytes with respect to redox potentials [6] and mitochondrial Ca2+ handling [13, 14]. However, these florescence sensors can be complicated by contaminating cytosolic signals. Previous cardiac myocyte studies have mostly focused on comparing isolated SSM and IFM [8–12], but less is known about how IFM and PNM differ in myocytes. Here we use real-time confocal imaging on live cardiac myocytes to examine morphological and functional differences between IFM and PNM.
In many cell types mitochondria are described as “highly dynamic”, including frequent mitochondrial fission/fusion events and translocation within cells [15–18]. Indeed, mitochondria in many cell types are dynamically rebuilt through continuous fusion and fission process in response to various cellular signals. Mitochondrial fission is required to create new mitochondria, but it also allows segregation of damaged mitochondria for degradation. Mitochondrial fusion produces elongated mitochondria and allows exchange of materials between mitochondria. Hence, mitochondrial fusion and fission appear to occur in a constant and balanced manner to allow the mitochondrial network to adapt to metabolic needs of the cell.
We hypothesize that in cardiac myocytes PNM exhibit dynamic fission/fusion and translocation similar to that described in other cell types, while IFM may be much more static, constrained in part by their rigid organization in a “lattice” of parallel rows surrounded by contractile myofilaments. The myofilament and Z-line structures could severely limit both movement and dynamic interaction between IFM. Despite these spatial restrictions cardiac IFM may exhibit dynamic interaction via nanotunnels and fusion/fission, for which the key proteins are all expressed in cardiac myocytes [19–21]. However, there is limited direct comparison of mitochondrial motility differences in IFM vs. PNM. Here, we use multiple approaches to examine the morphology and functional dynamics of IFM vs. PNM in adult cardiac myocytes. Our data indicated that IFM and PNM vary substantially in morphology, Ca2+ uptake, permeability transition pore (PTP) opening, motility, fission/fusion events and mitochondrial turnover via mitophagy.
Results
Mitochondrial distribution and morphology (IFM vs. PNM)
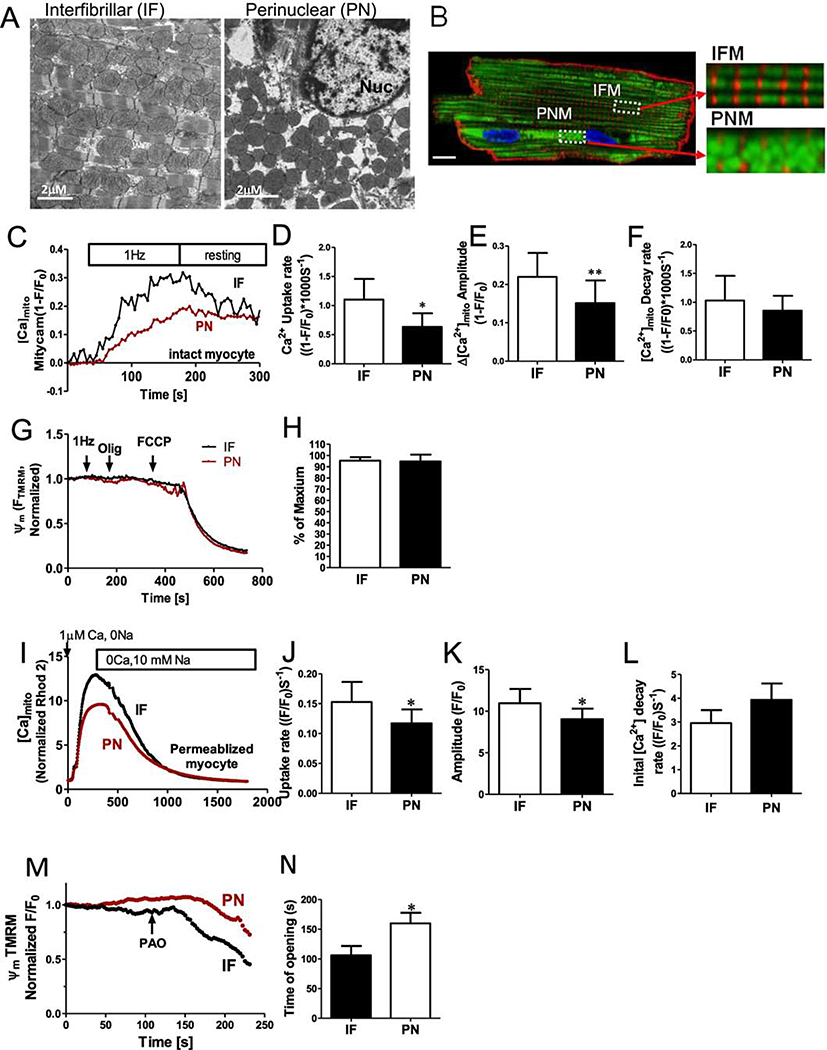
We studied mitochondrial organization and function using confocal imaging and electron microscopy (EM) of intact and permeabilized rabbit ventricular myocyte. As classically described in EM images [22] and in Figure 1A, IFM in adult ventricular myocytes are aligned in longitudinal chains between the myofibrils, and seem to be squeezed into these tracks by the myofilaments which perforce are longitudinally continuous. This may contribute to the typically oval shape of IFM, and further lateral compression would be expected during contraction as the sarcomere thickens. In contrast PNM distribute around the nucleus and in the typically binucleate myocyte also in the space between the two nuclei. These PNM typically appear more spherical in shape (Figure 1A). To quantify mitochondrial morphology in these EM images, mitochondrial outer perimeters were individually traced. The major mitochondrial shape descriptors including perimeter (mean±SD for IFM vs PNM respectively: 3.96±0.16 vs 3.0±0.13 μm, p< 0.001), aspect ratio (1.89±0.08 vs 1.40±0.03, p< 0.001), circularity (0.757±0.012 vs 0.825±0.008, p< 0.001), and form factor (FF) (1.38±0.03 vs. 1.24±0.02, p< 0.001) differed significantly between IFM and PNM (Supplemental Fig1). Circularity is a two-dimensional index of sphericity (4π• (surface area)/(perimeter)2) where 1.0 indicates a perfect circle (or sphere). PNM are smaller, more spherical and exhibit shorter perimeter, whereas IFM have larger aspect ratios. Notably, PNM do lack the myofilament constraints and that may contribute to the more spherical shape of PNM. (Supplemental Figure1A–C). FF (the inverse of sphericity) also reflects complexity and branching aspects of mitochondria.
Figure 1. Perinuclear and intermyofibrillar mitochondrial morphology and Ca2+ uptake in intact cardiac myocytes.
(A) Electron microscopy images of sections through left ventricular rabbit heart. Intermyofibrillar mitochondria (IF) are often a rounded brick-shape (left), and perinuclear mitochondria (PN) are more round and loosely arranged (right). (B) Ad-Mitycam indicates mitochondria (and [Ca]mito), Di-8-ANEPPS indicates T-tubule (red) and Hoechst 33342 indicates the nucleus. Enlarged images from the indicated myocyte regions in B (scale bar 10 μm). (C) Kinetics of [Ca]mito during 1 Hz pacing frequency in adult rabbit cardiac myocytes, mean initial [Ca]mito uptake rate (D), Δ[Ca]mito amplitude (E) and decline rate[Ca]mito when pacing stopped (F; n=6 cells). (G,H) Mitochondrial membrane potential Δψm after 1 Hz pacing, normalized to the maximal value with oligomycin (n=6 cells). Kinetics of [Ca]mito change during Ca2+-clamp in permeablized (and SR disabled) cardiac myoyctes (I), mean Δ[Ca]mito (K), initial uptake rate (J) and [Ca]mito decline rate (L; n=6 cells). M,N ψm was normalized to the initial value, and mPTP opening time induced by phenylarsine oxide (PAO; n=4 cells) was estimated as the time when a regression line for the first 20 points of sustained ψm decay intersect the baseline.
Mitochondrial Ca2+ handling in IFN vs. PNM
We measured mitochondrial Ca2+ uptake in IFN vs. PNM in both intact and saponin-permeabilized myocytes. Mitochondrial Ca2+ participates in excitation–metabolism coupling. That is, when Ca2+ transients are larger or more frequent mitochondria accumulate more Ca2+, which enhances ATP production via Ca2+-dependent dehydrogenases during oxidative phosphorylation [23–25]. First, we measured mitochondrial Ca2+ uptake caused by electrical field stimulation of intact myocyte (at 1 Hz) after a long period of rest in low [Ca2+] containing solution. The genetically encoded and mitochondrially targeted Ca2+ sensor Mitycam was expressed via adenoviral infection as previously described [5]. The Mitycam signal indicates that intra-matrix free [Ca2+] ([Ca2+]Mito) gradually rises during pacing over 2–3 min until pacing is switched off, allowing slow [Ca2+]Mito decline. IFM exhibited much faster initial rate of Ca2+ uptake (1.10±0.35 vs. 0.63±0.23 s, *p<0.05) and larger amplitude (0.220±0.06 vs. 0.151± 0.06, *p<0.05, Figure 1 C–E). However, the post-stimulation [Ca2+]Mito decline, while faster in the IFM example in Fig 1C, was not significantly different on average between IFM and PNM (Figure 1F). Figure 1G shows measurement of mitochondrial membrane potential (ψm) which was close to the negative maximum achieved by inhibition of the F0F1-ATP synthase (typically–180 mV) in both IFM and PNM, even during the period of 1 Hz myocyte stimulation. No difference in ψm was detected between IFM and PNM.
The faster Ca2+ uptake observed in IFM could be due to an intrinsic differences in MCU-dependent uptake rate, but in intact myocytes, local SR Ca2+ release could create higher [Ca2+]i around IFM vs. PNM [5]. To better assess the intrinsic mitochondrial Ca2+ uptake that is independent of SR Ca2+ release, we also measured mitochondrial Ca2+ uptake in saponin-permeablized myocyte with 5 μM thapsigargin to prevent SR Ca2+ uptake and release, and with [Ca2+]i clamped by heavy buffering we raised [Ca2+]i in a step from 0 to 1 μM (Figure 1I). IFM still exhibited faster initial Ca2+ uptake rate vs. PNM (0.153±0.03 vs. 0.117± 0.020, *p<0.05), and larger amplitude (10.96±1.73 vs. 9.03±1.28, *p<0.05 Figure 1I–J), but no difference in the rate [Ca2+]Mito decline (Figure 1L). However, the IFM : PNM uptake rate ratio was smaller in permeabilized vs. intact cells, consistent with the idea that some intrinsic differences (present in the permeabilized case) are further amplified by local SR Ca2+ release associated with excitation-contraction (E-C) coupling in intact myocytes.
We also tested whether IFM or PNM were differentially sensitive to oxidative stress induced by exposure to phenylarsine oxide (PAO) application (Figure 1M–N). We measured ψm in permeabilized myocytes (which was not different at baseline), but found that the IFM were more readily depolarized by PAO. This might reflect either higher oxidative status or a higher [Ca2+]m in IFM, which would favor mPTP opening. Taken together, these findings indicate that IFM and PNM have different Ca2+handling properties and sensitivity to oxidative stress.
Mobility of mitochondria in adult cardiac myocytes (PNM vs. IFM)
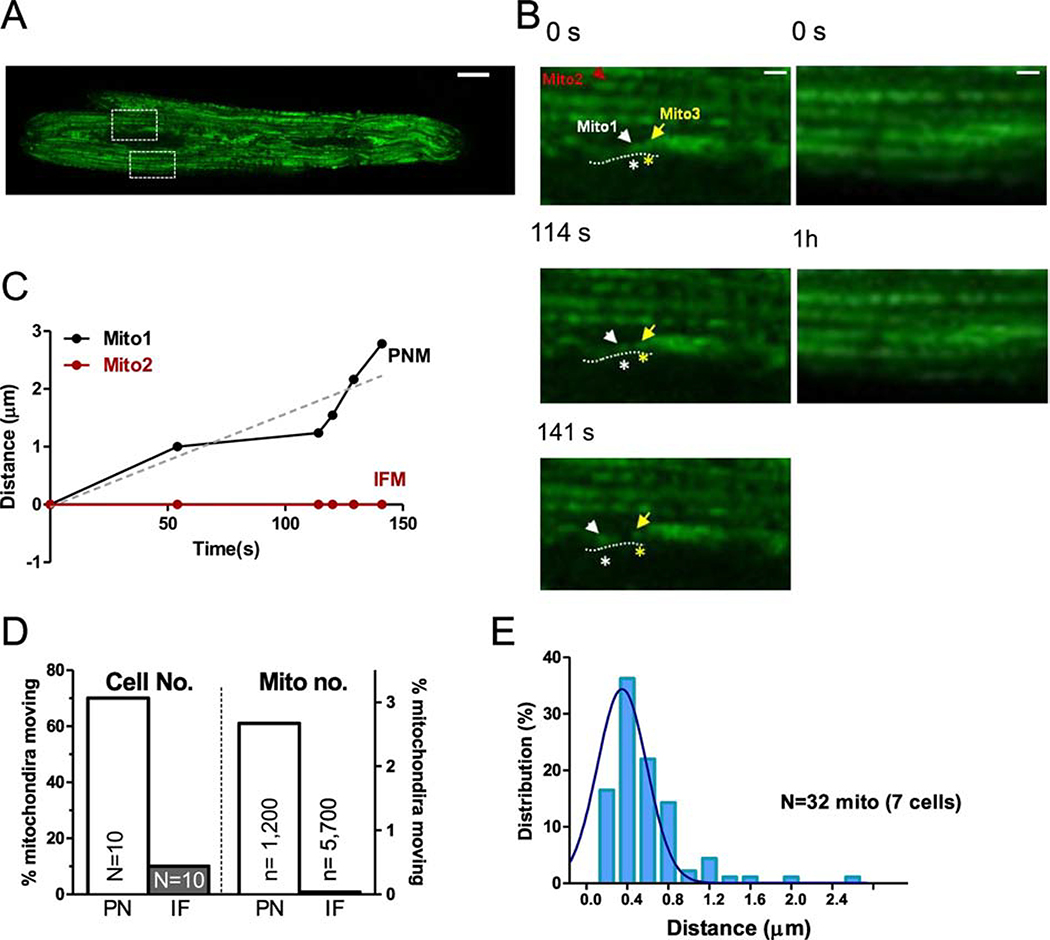
Mitochondria are very dynamic in cells lacking the rigid structure of cardiac myocytes. Here we tracked the movement of individual PNM vs. IFM using mitochondrially targeted GFP (after Ad-mtGFP expression for 24–36 hr) via confocal imaging of live cardiac myocytes in real time. During a 1 hr observation period, IFM essentially did not move (Figure 2A–C). Only 0.04% of IFM moved detectably (2 IFM out of ~5700 mitochondria in 10 cells) and the net movement of the mitochondrion in Figure 2D was less than 0.3 μm. For PNM we observed movement in 70% of the 10 myocytes imaged during 1 hr of observation (and 32 out of ~1200 total PNM followed). An individual PNM tracked with mtGFP in Figure 2B–C (mito1) traveled 2.8 μm along the indicated path during 140 s before stopping, in contrast to a simultaneously monitored IFM (mito2). The average distance of mobile and total PNM (including the static PNM) was ~0.53 and 0.01 μm respectively (Figure 2E).
Figure 2. Motility of individual mitochondrion in adult cardiac myocytes.
(A) Right: expression of mitochondrial indicator Ad-mtGFP in adult cardiac myocytes (scale bar: 10 μm), (B) Enlarged images from the indicated myocytes regions in A (IF: lower ROI and PN: upper ROI) at different time points (scale bar: 4 μm). (C) Distance moved by individual mitochondria indicated in B (IFM: mito2 and PNM: mito1) Mito1 (labeled with white *) moved along the nuclear edge indicated by dash lines and away from the neighbor mitochondrion Mito3 labeled with yellow *. (D) Percent of cells (N, left)) and mitochondria (n, right) exhibiting movement and (E) distribution of distances moved by individual mitochondrion moved in PNM and IFM (n= 7 cells).
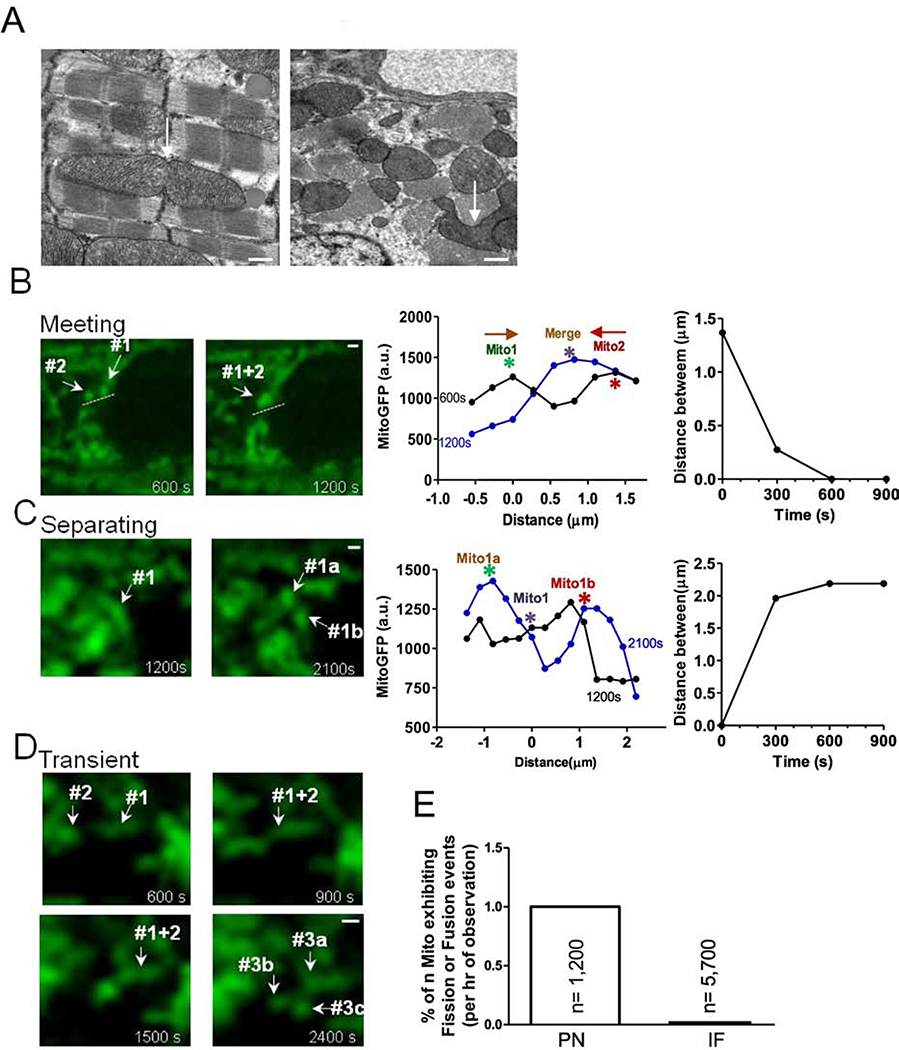
Mitochondria can also participate in fusion and fission. In static EM images we found some branched, potentially transitional-shaped mitochondria, which might be undergoing fusion or fission (Figure 3A). To assess this possibility dynamically we used Ad-mtGFP infected adult cardiac myocytes to test whether fusion/fission events could be visualized in live adult myocytes. Figure 3B shows two individual PNM that start 1.4 μm apart and move toward each other, apparently fusing into one mitochondrion. We also observed mitochondria which appear to split or separate (Figure 3C) and others that seem to transiently interact (Figure 3D). These events were almost exclusively observed in PNM rather than IFM (Figure 3E).
Figure 3. Visualization of mitochondrial interaction in adult cardiac myocytes.
(A) EM images showed mitochondrial shapes (scale bars: 500 nm). (B) Two mitochondria move towards each other along the path indicated by dash lines. Right: position and distance of mitochondrion center indicated in left image at different time points. (C) mother mitochondrion separates into two daughter mitochondria. Right: position and distance of mitochondrion indicated in left image at different time points. (D) Transient mitochondrial fusion (scale bars: 2μm). (E) Percent of mitochondria exhibiting dynamic interaction.
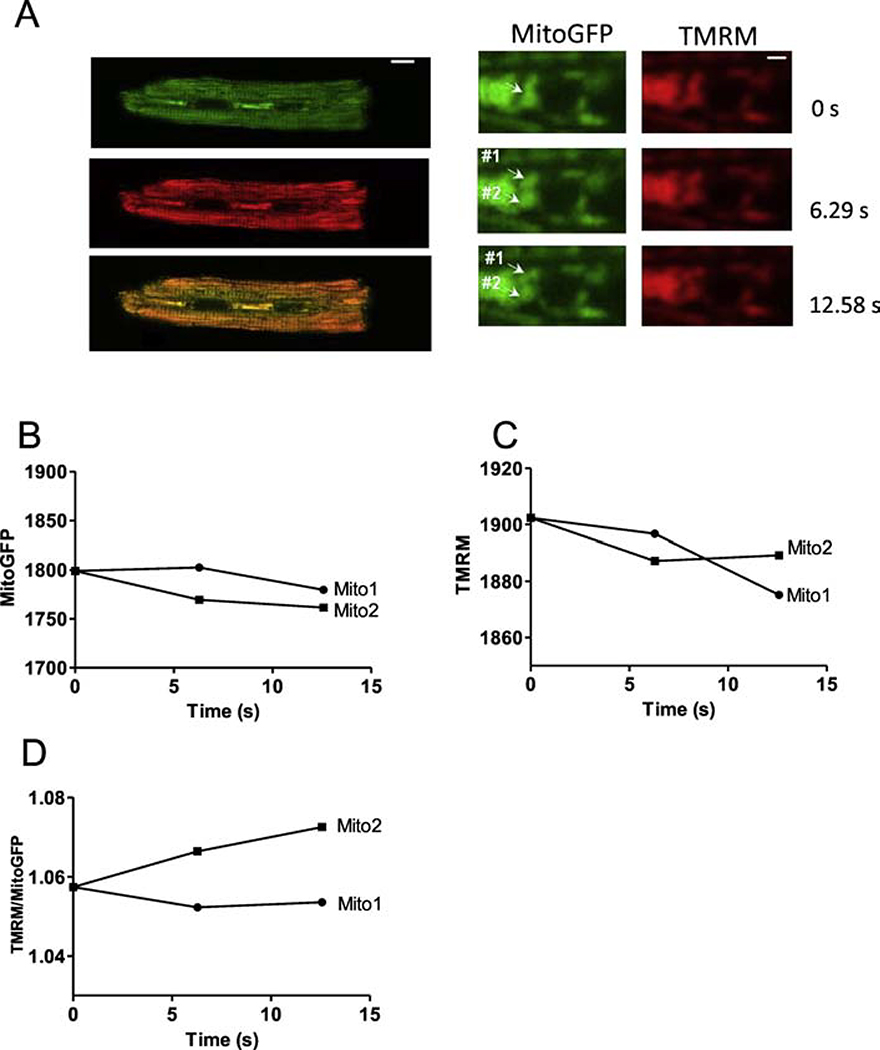
To further test whether such apparent fission/fusion events are associated with mitochondrial depolarization (Δψm) we monitored TMRM and mtGFP in Ad-mtGFP transfected cardiac myoctyes. Most of the events observed showed unaltered Δψm before and after fission/fusion. However, two events were like Figure 4, in which one apparent daughter mitochondrion (mito2) increased ψm (normalized to mtGFP) vs. the other daughter (mito1) slightly decreased ψm after normalized to mtGFP (Figure 4D). This apparent differential ψm health in daughter mitochondria resembles results reported in COS7 cells [17].
Figure 4. IMM Δψm during mitochondrial separation.
(A) Cardiac myocytes expressing Ad-mitoGFP (green) and mitochondrial membrane potential sensor TMRM (red) (scale bar: 10 μm). Right: enlarged images from the indicated myocytes region in left images at different time points (scale bar: 2 μm). Fluorescence of mitoGFP (B),TMRM (C) and the TMRM/ mitoGFP ratio (D) of mito1 and mito2 (in A) as a function of time.
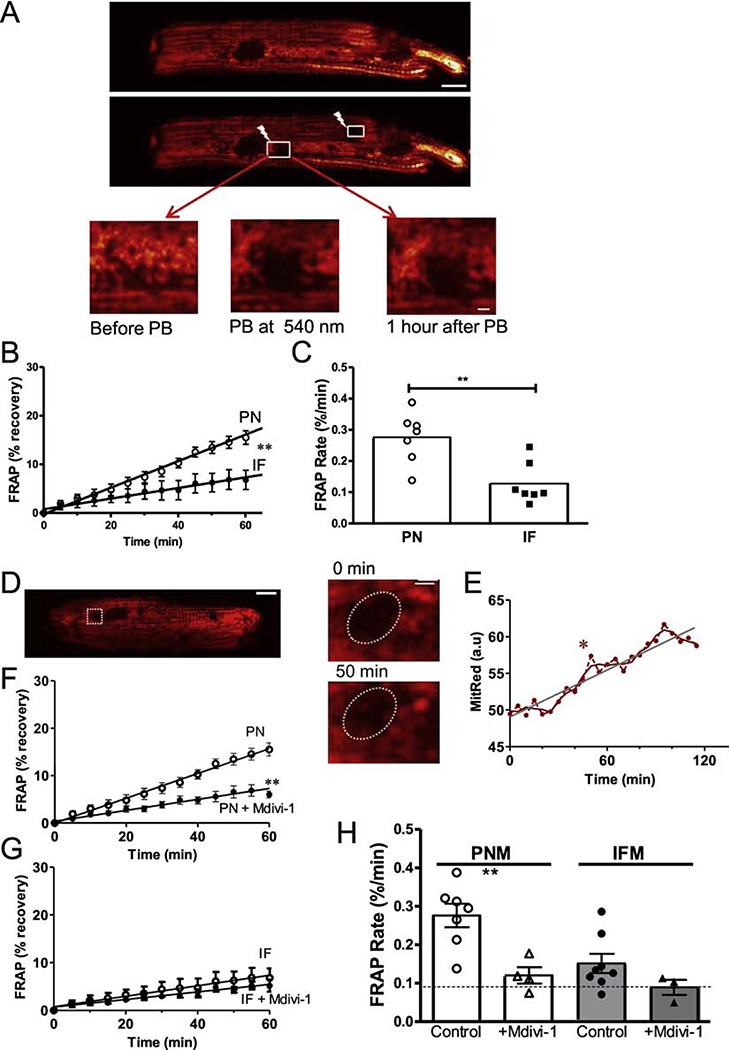
To further test whether mitochondrial matrix content is exchanged in adult cardiac myocytes, we expressed mitochondrial matrix-targeted Ad-mtDsRed in cultured cardiac myocytes and measured fluorescence recovery after photo-bleaching (FRAP). If FRAP occurs in an individual mitochondrion that would indicate mixture of contents with an unbleached mitochondrion. But in the FRAP region, an unbleached mitochondrion could also move into that region (reflecting motility). We compared the FRAP recovery rate for IFM and PNM for up to 1 hr after photobleach in that given cell. PNM showed much faster FRAP rates vs. IFM (0.273 ± 0.014 vs. 0.110 ± 0.018 %/min, p< 0.01, Figure 5A–C). The higher PNM motility would also affect the FRAP rate (in addition to fusion/fission). Indeed, the FRAP shown in Figure D-E shows one mitochondrion that moved into the oval FRAP region (see enlarged 50 min image; Figure 5D). This was responsible for the rapid increase of the florescence at 50 min (labeled with *), which is much faster than the average FRAP rate (Figure 5E). Thus, both mitochondrial fusion/fission and motility contribute to the FRAP in the PMN region. To more specifically assess the contribution of fusion/fission to the observed FRAP rate, we suppressed fusion/fission using the Drp-1 inhibitor mdivi-1 [26, 27] (that can also partially inhibit Complex I [28]). Mdivi-1 strongly suppressed FRAP rate in PNM, but not in IFM (Figure 5F–H). This suggests that more than half of the FRAP in PMN was due to fission/ fusion events, with mobility being responsible for less than half. The extremely low IFM mobility directly monitored in Figure 2 might suggest that IFM FRAP with mdivi-1 (0.089%/min; dotted line) is an effective technical baseline, and if so fission/fusion may exceed mobility for both PMN and IFM FRAP.
Figure 5. PNM have faster fission/fusion rates than IFM.
(A, D) Confocal images of cardiomyocytes transfected with Ad-DsRed before and after photobleaching (PB) the ROIs at 540 nm (scale bar: 10 μm, scale bar of enlarged images: 2 μm). (B, C) FRAP time course and rate for PN and IF mitochondria. (D,E) Exemplar ROI where an existing mitochondrion appears to enter the ROI during FRAP period. (F-H) FRAP of DsRed signal after photobleaching with and without Drp1 inhibition by Mdivi-1 (n=7 cells).
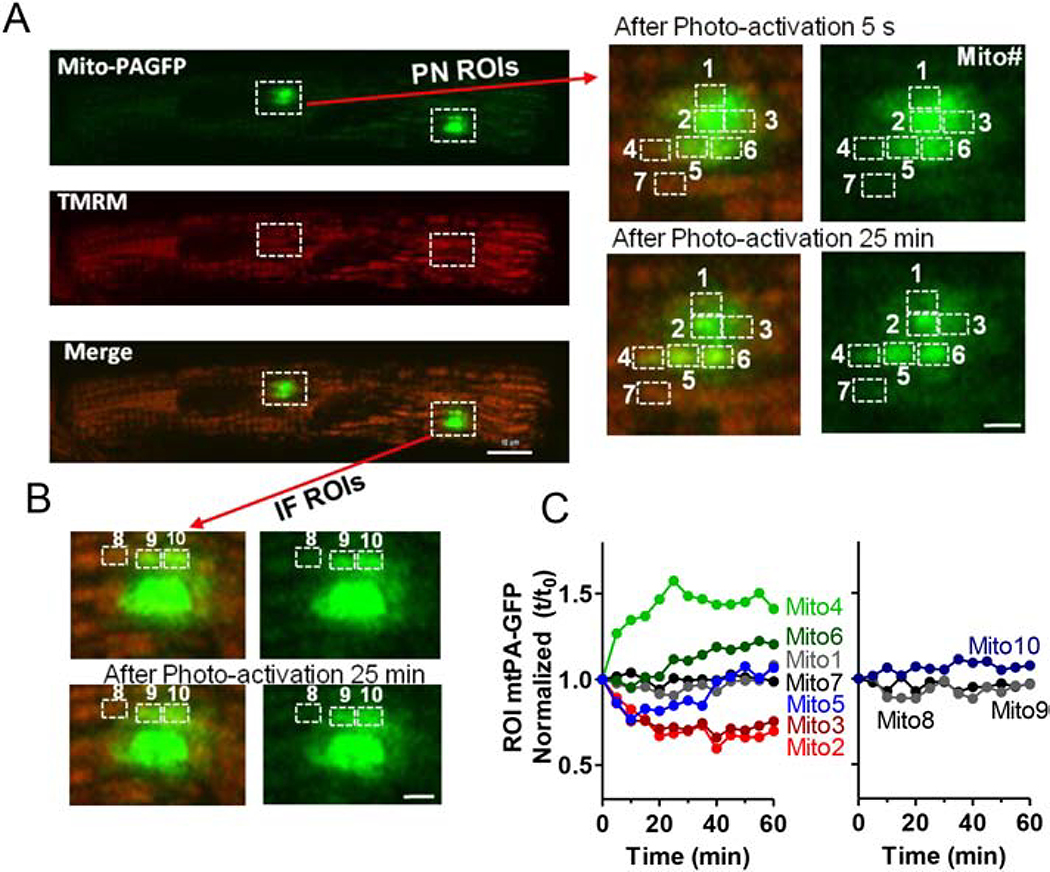
In addition to these FRAP experiments, we also used mitochondria-targeted and photoactivatable GFP (mtPA-GFP) to produce GFP fluorescence only in a small cluster of mitochondria whose fate could be tracked. Myocytes expressing mtPA-GFP (via adenovirus) were also labeled with TMRM to identify all mitochondria. After photoactivation of mtPA-GFP at 405 nm in PNM and IFM regions, we monitored fluorescence at 488 nm (Figure 6A, left). The enlarged PNM region of interest (ROI; Figure 6A, right) show abrupt photo-activation in several local mitochondria. In Mito2 and 3 (in the photoactivated region) fluorescence progressively declined after photo-activation (by ~35% over 25 min; Figure 6C) while a neighboring mitochondrion (Mito4) just outside of the photo-activated region exhibited a 50% rise in fluorescence (from 69 to 103). Mito6 just below Mito2–3 also showed increased GFP, while Mito5 first lost and then gained GFP fluorescence. In contrast, Mito7 (located just below Mito4) fluorescence did not change significantly. This is consistent with Mito2, 3 and 5 transferring some GFP to Mito4 and 6, with Mito5 getting some back later from Mito2–3. This apparent matrix mixing between individual PNM presumably results from fission/fusion events. Analogous analysis of an IFM ROI in this same myocyte did not exhibit significant fluorescence shifts after photo-activation (Mito8–10; Figure 6B–C).
Figure 6. Visualization of intermitochondrial communication in adult mouse cardiac myocytes.
(A) Confocal images of cardiomyocytes transfected with Ad-mtPAFGP (Green) and stained with TMRM (Red) indicating individual mitochondrion, at different time points after photoactivation of indicated ROIs. Right: Enlarged PNM region indicated in left images. (B) Enlarged IFM regions indicated in (A) (scale bars: 4 μm). Time courses of mtPA-GFP fluorescence signal of PN and IF mitochondria over time. Individual mitochondria were numbered in A and B as indicated to identify fluorescence changes in each mitochondrion (C).
To test whether this apparent PNM vs. IFM difference in mitochondrial dynamics is related to distribution of fusion/fission machinery, we immunostained for the fission protein Drp1, and fusion proteins Mfn1, Mfn2, and OPA, and co-stained with MitoTracker to identify mitochondria. We did not detect striking differential localization (PNM vs. IFM) of these mitochondrial dynamic proteins (Supplement Figure 2).
Perinuclear translocation may be critical in Mitochondrial Turnover
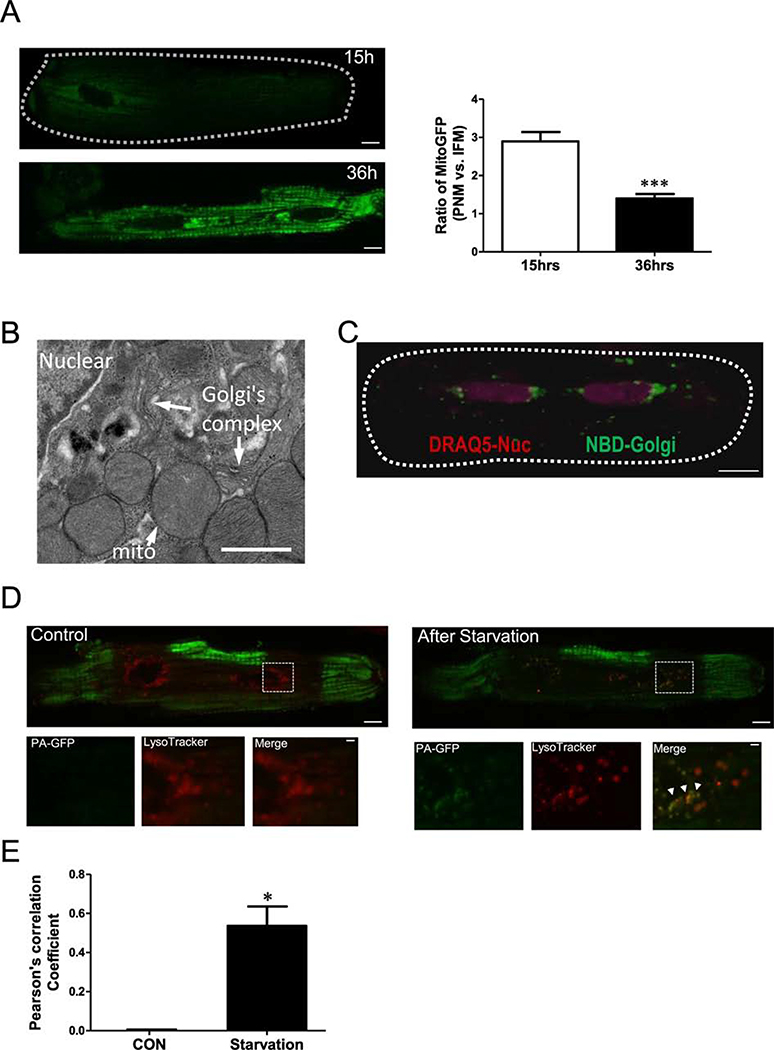
Unlike other cell types, adult cardiac myocyte mitochondrial trafficking is complicated by myofilament structural constraints. Here, we explore the idea that mitochondrial biogenesis initiates preferentially in the perinuclear region. We transfected Ad-mtGFP into cardiac myocytes and monitored mtGFP expression. Fifteen hours post-transfection, mtGFP was highly expressed in PNM, but less so elsewhere, while at 36 h post-transfection mtGFP expression was more uniform throughout the myocyte (Figure 7A). While this might be due to mitochondrial genesis, it might only reflect the faster protein translation in the perinuclear area (and hence accumulation by local PNM). Our EM data does show perinuclear Golgi enrichment near these PNM (Figure 7B). In live myocytes, confocal imaging using DRAQ5 to stain nuclei and NBD to indicate Golgi, we also find Golgi mainly in the perinuclear region (Figure 7C). Thus, PNM are physically closer to protein synthesis sites for perinuclear mitochondrial biogenesis.
Figure 7. Perinuclear region is highly active site for starvation-induced mitochondrial turnover.
(A) Confocal images of mitochondrial GFP expression in cardiomyocytes at 15 and 36 hrs after transfecting Ad-mitoGFP. (B) EM image, (C) NBD Golgi staining in live cells indicated that Golgi is enriched around nuclear (indicated by DRAQ5, violet). (D) Confocal images of cardiomyocytes transfected with Ad-mtPA-GFPafter photoactivating ROIs (green) at the cell periphery and stained with LysoTracker red to indicate lysosomes (Red), lower panels are enlarged images from the indicated myocytes region in upper panels. (E) Pearson’s correlation coefficient of mtPA-GFP and LysoTracker signal.
This raises a question as to whether the perinuclear region is also related to mitochondrial clearance and turnover via mitophagy. To encourage mitochondrial turnover via starvation, we cultured cardiac myocytes in glucose free starvation media for 6 hr [29]. Starvation can trigger mitophagy, where unhealthy mitochondria are degraded in autophagosomes and lysosomes. We measured mitochondria and lysosomal co-localization as an index of where mitochondrial turnover is occurring in live myocytes. Individual mitochondria were visualized using mtPA-GFP (green) and lysosomes were labeled using LysoTracker (red; Figure 7D). We photoactivated mtPA-GFP in multiple intrafibrillar regions before starvation (green in Figure 7D, and observed mainly perinuclear lysosomes (red) at that time. Six hours after starvation, mtPA-GFP from IFM was found to co-localize with lysosomes in the perinuclear region (Figure 7D, right). This suggests that unhealthy mitochondria (or fission products therefrom) were delivered from myofilaments or subsarcolemmal regions to the perinuclear area where they fuse with lysosomes to be degraded (Pearson’s correlation: 0 vs. 0.5360±0.1, P<0.05, Figure 7E).
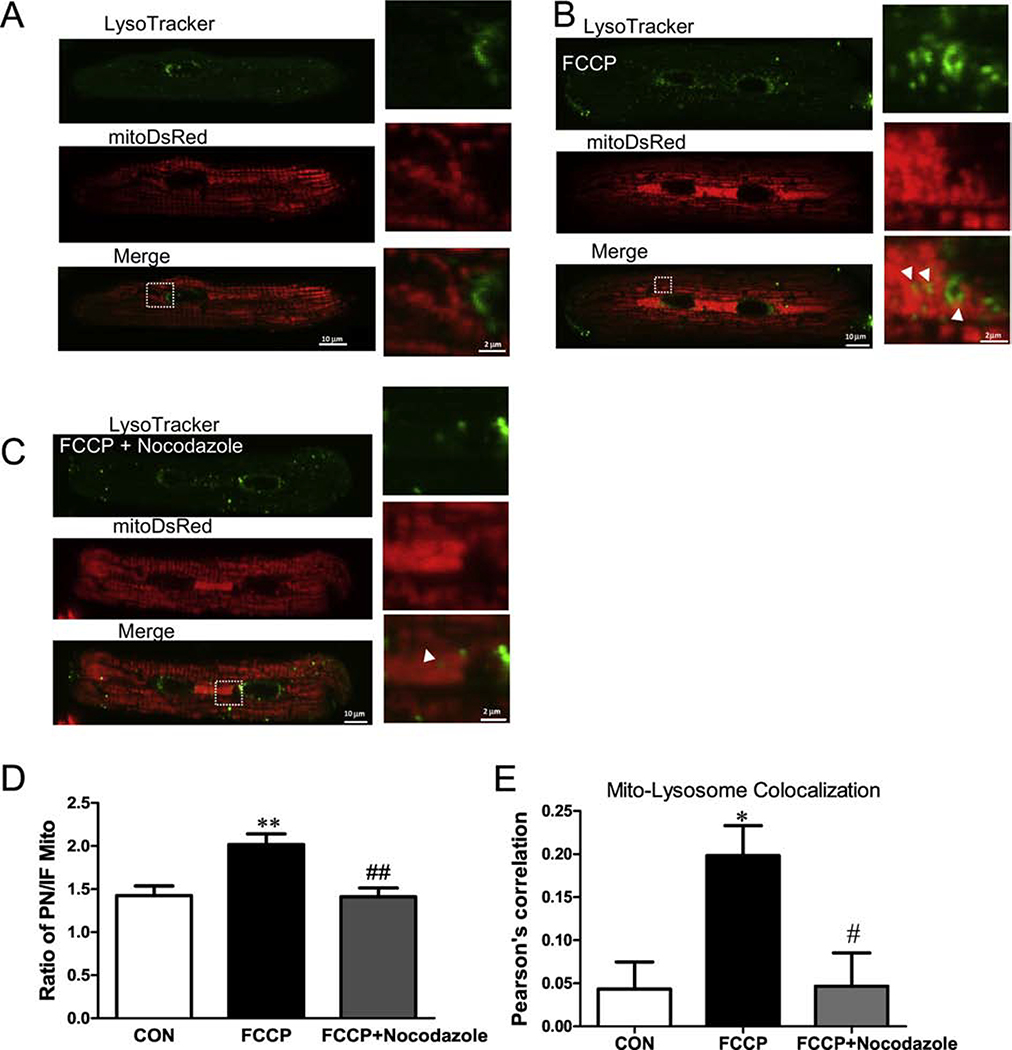
Next, we induced mitophagy by exposing myocytes to a mitochondrial depolarizing stress (100 nM FCCP overnight; Figure 8). Here, myocytes expressed mtDsRed to label individual mitochondria and we used LysoTracker green to track lysosomes. Our data showed little evidence for mitophagy under normal conditions, but after a 6h FCCP exposure there was increased co-localization of mtDsRed and LysoTracker (Pearson’s correlation: 0.04± 0.03 vs. 0.20± 0.03, **P< 0.01, Figure 8A, B, E). Moreover, we also observed an increased perinuclear mtDsRed intensity after FCCP application. This might reflect delivery of unhealthy mitochondria from intrafibrillar to the perinuclear region. To further test this idea, we applied 5 μM Nocodazole to disrupt microtubule assembly. Nocodazole significantly inhibited translocation of mitochondria to the perinuclear region (reduced PN/IFM mtDsRed; Figure 8C–D) and the co-localization of mitochondria with lysosomes (Pearson’s correlation: 0.20± 0.03 vs. 0.05± 0.03, ##P< 0.05, Figure 8C, E).
Figure 8. Inhibition of mitochondrial trafficking to the lysosome at the perinuclear region.
Cardiac mycoytes stained with LysoTracker green and expressing mtDsRed before (A) and after (B) applying 100 nM FCCP for 6 hr. (C) Ratio of DsRed signal (PNM/IFM) with and without microtubule disruption (Nocodazole) during FCCP treatment. (D) Pearson’ correlation coefficient of mito-DsRed and LysoTracker green with and without Nocodazole during FCCP treatment (n=6–12 cells).
We conclude that IFM are normally non-motile with little basal fission/fusion activity (in contrast to PNM). However, upon mitochondrial damage, IFM (or their fission fragments) make their way toward the perinuclear region via microtubules to participate in lysosomal mediated mitophagy.
Discussion
Mitochondria are crucial determinants in the life and death of cells and changes in their morphology and function underlie processes [30, 31]. Unlike many cell types, in cardiac myocytes the rigid myofilament and Z-line structures may cause morphological difference between IFM and PNM. The proximity of IFM to the intense energy demands placed on the myofilaments may also contribute to functional differences in mitochondrial Ca2+handling. Here, we compared these two mitochondrial sub-populations from different perspectives and investigated the functional significance of their regional differences.
Mitochondrial Structure and Function are Tuned to Subcellular Locations
Cardiac muscle requires large amounts of mitochondrially produced ATP for energetic demands. Heterogeneity of mitochondrial oxidative differences were first demonstrated by Palmer et al. [10] using isolated mitochondria from SSM and IFM subsets. Several studies have reported that IFM (vs. SSM) exhibited 50% higher levels of substrate oxidation and higher activity of key oxidative phosphorylation enzymes, including succinate dehydrogenase and citrate synthase [10, 32]. This difference might be the result of the specific physiological roles reflecting different subcellular niches. The IFM are thought to generate most of the ATP for contraction, while SSM may also provide energy supply for active sarcolemmal transport of electrolytes and metabolites.
In contrast, PNM are less physically constrained, allowing them to assume a more spherical shape and perinuclear clustering. PNM have been suggested to produce ATP close to the nucleus and influence nuclear function. In both intestinal Cajal-like cells and atrial cardiac myocytes, PNM play be critical in regulating local Ca2+ waves and SR/ER Ca2+ release frequency by buffering Ca2+ near the nucleus [33, 34]. Thus, spatially-defined mitochondrial subpopulations can sense and modulate their local energy supply and demand as well as Ca2+ signaling. This may also reflect distinct chemical and metabolic properties of spatially different mitochondrial populations [10, 35]. PNM may also regulate various nuclear functions [36], including modification of promoters to alter transcriptional complex assembly and mRNA expression [37].
Scanning EM showed distinct morphological cristae structure more dense in IFM vs. SSM, consistent with higher ATP production in IFM [11]. We also observed morphological differences between IFM and PNM (although we did not analyze cristae structure). IFM had larger perimeters, areas and lengths, with aspect ratios close to two (1.9) vs. the more circular PNM. Morphologically, PNM were for the most part isolated circular units with many loosely clustered around the nuclear poles. Our data is 2D EM images, so we do not know if Z-direction depths would match the shorter 2D dimension, but our results are consistent with a previous study using 3D EM images [38]. IMF mitochondria are organized into a lattice-like string of elongated beads alongside the densely packed force bearing myofibrils, with each mitochondrion typically spanning the length of one sarcomere, bounded longitudinally by dense protein structures at Z-lines (~1.8 μm; Figure 1).
As above, the IFM may be specialized to provide high levels of ATP to support local myofilament energy demands, such that mitochondrial number and morphology are influenced by the local energetic cellular requirements [39]. Hence, in adult cardiac myocytes, the larger and more active IFM (vs. PNM) could be an adaptation to the higher energetic demands. This is also functionally in-line with our observation of higher IFM Ca2+ uptake in intact cells, which can promote oxidative phosphorylation and ATP synthesis.
Here, we measured [Ca2+]Mito rise during both pacing and Ca2+ transients in intact myocytes and in response to a [Ca2+]-clamp pulse to 1 μM in permeabilized myocytes with the SR disabled (preventing SR Ca2+ release induced spatial gradients). In both cases, the initial rate of [Ca]Mito rise (Fig 1D and J) and peak of Ca2+ uptake were higher in IFM than PNM. This difference was apparent in the Ca2+-clamp experiments suggesting that there are intrinsic differences between IFM and PNM in situ. The fact that this difference was more dramatic during physiological Ca2+ transients, suggests that the IFM (vs. PNM) also have some degree of preferential access to Ca2+ released from the SR during E-C coupling. There is direct experimental evidence for higher and faster increases of [Ca2+]Mito near the location where SR Ca2+ release occurs vs. other locations (mid-sarcomere and perinuclear region) which experience a [Ca]i signal closer to the cytosolic average [5].
The organization of IFM in cardiac myocytes also allows diffusible cytosolic messengers to promote propagating waves of mitochondrial depolarization that can oscillate rhythmically and cause cyclical changes in overall cellular energetics [40–42]. Notably, these oscillations do not require direct physical interaction of neighboring mitochondria discussed below, but are driven by diffusion of reactive oxygen species (ROS) via ROS-induced ROS release waves [7, 43]. Hence, cytosolic diffusible messengers, local architecture, and the intrinsic mitochondrial differences could all contribute to rapid and sensitive response to changes in energy demand, but can bring with it the potential for metabolic instability.
Mitochondrial Movement, Fusion and Fission
We monitored mitochondrial movement by tracking individual mitochondrial position over time (Figures 2–3), FRAP of mitochondrial targeted DsRed (Figure 5 and 8) and photoactivation of mtPA-GFP (Figure 6–7). In these techniques movement can be complicated by exchange of mitochondrial content mediated by fission/fusion events and direct nanotunnels connecting nearby mitochondrial matrices [19, 20].
Most cells lack the rigid mechanical structure that surrounds IFM in cardiac myocytes. So it is was not surprising that we found IFM to be nearly fixed in position for times up to 1 hour in ventricular myocytes, which contrasts to substantial mobility of PNM (Figures 2, 3, 5 and 6). The exemplar PNM in Figure 2B–C moved along the nuclear edge, suggestive of directed rather than random movement. That is consistent with strong evidence in neurons that mitochondria are transported along microtubules over long distances [44], and that when we disrupted microtubules with nocodazole mitochondria were inhibited from traveling to the perinuclear region during mitophagy (Figure 8). Thus, while random movement in the less confined perinuclear region may occur, much of the directed mitochondrial translocation in cardiac myocytes may be along the rich microtubule network in these cells including the perinuclear region [45, 46].
The FRAP experiments illustrated best how both movement of unbleached individual mitochondria into the bleached region may explain nearly half of the FRAP measured in PNM. That is implied by the fact that inhibiting fission/fusion reduced FRAP by >50% (Figure 5F). The IFM FRAP was much slower, and inhibition of fission/fusion had a negligible effect (Figure 5G). Since there was little evidence for IFM movement in this time frame, a substantial part of the very slow IFM FRAP (with fission/fusion inhibited) might be mediated by the relatively stable nanotunnels described by Huang et al. [19]. That is, even without clear fission/fusion events, exchange of mitochondrial content between neighboring mitochondria can occur [19, 20] and that may be the dominant pathway for IFM, while fission/fusion and mitochondrial movement all contribute importantly to PNM FRAP. Eisner et al. found that cultured vs. fresh cells exhibited different mitochondrial dynamics, so we cannot rule out the possibility that IFM and PNM respond differently to culture that was required for sensor adenoviral transfection [20]. However, we did not observe different responses to culture between PNM and IFM, and our Rhod-2 experiments using fresh isolated cardiac myocytes gave data that were consistent with the Mitycam [Ca]Mito measurements.
Mitochondrial fission/fusion also appears to differ dramatically between PNM and IFM. PNM exhibited much more apparent fusion and fission activity vs IFM. Fusion-mediated exchange of soluble matrix mtPA-GFP was readily observed in PNM, but not in IFM during an hour of observation (Figure 6). In addition, the mdivi-1 sensitivity of FRAP was large for PNM, but not in IFM, indicative of greater fission/fusion in PNM vs. IFM. This could be partly caused by limited motion of IFM as observed experimentally and physical barriers between neighboring IFM (Z-lines longitudinally and sarcomeres transversely). Despite this, the apparent distribution of fusion/fission proteins was similar in IFM and PNM Suppl Figure S2). Thus, the machinery is present (and that may be required for IFM turnover discussed below). Of course, other factors may limit the local activities of these fission/fusion proteins (e.g. recruitment to the mitochondria, exact locations with respect to regulatory protein partners, GTPases and post-translational modifications). Thus, while there is abundant expression of these fusion/fission proteins, the fission/fusion activity could be highly regulated in cardiac myocytes. We may underestimate the absolute frequency of fission/fusion events in intact hearts because we studied primary myocytes 24–30 hr after adenoviral infection in culture to reach adequate expression of the fluorescent proteins, which Eisner et al. [20] found to decrease these events. However, we do not think this is likely to alter our comparisons between IFM and PNM in the same cells.
Mitochondrial Turnover and Mitophagy
Mitochondria have tissue-specific rates of turnover. In mouse heart, mitochondria turn over with a half-life of 14 days [47], but in the liver, the half-life is only 2–4 days[48, 49]. Selective mitochondrial autophagy, or mitophagy, eliminates damaged and dysfunctional mitochondria [48] and is closely linked to mitochondrial biogenesis, which permits replacement with healthy mitochondria and. Baseline mitochondrial turnover in healthy myocytes is slow enough to make it hard to study in single cell/mitochondrial studies. So, we stressed myocytes by starvation or FCCP to make the process more practical to monitor.
We observed starvation-induced mitophagy in the perinuclear space, and that IFM (with photo-activated mt-PA-GFPh) had moved there from the interfibrillar space to undergo mitophagy (Figure 7). FCCP also promoted mitochondrial translocation to the perinuclear region and co-localization with Lyso-tracker. Lysosomes are relatively concentrated in the perinuclear region. The less structured space and mitochondrial delivery there may facilitate mitophagy. Moreover, microtubular disruption with nocodazole prevented the net translocation of mitochondria to the perinuclear space, and the extent of mitochondrial-Lyso-Tracker colocalization (Figure 8). Thus, the perinuclear region may be the active site for mitochondrial clearance (for both IFM and PNM) and microtubules are important in delivering damaged IFM mitochondria to the perinuclear region to undergo mitophagy. The signaling pathways involved in mitophagy are complex and multifold [50, 51], but the spatial migration of mitochondria as described here may be an important part of the process. Future studies will be required to determine at what exact stage (and molecular steps) in the mitophagic pathway lead to microtubule-mediated translocation of IFM to the perinuclear myocyte domain.
Golgi are also enriched around the nucleus, which could also favor mitochondrial biogenesis. Our data showed earlier mitochondrial GFP (mt-GFP) expressed in PNM compared to IFM after transfection with mt-GFP. This suggests the IFM subpopulation displayed a lower protein synthesis rate than the PNM, or that mitochondrially targeted proteins are preferentially made near the nucleus and delivered more slowly to distant IFM. A previous proteomics study, showed cardiac protein synthesis is slower at IFM than SSM [9]. In neurons, it is thought that mitochondria are synthesized in the cell body and then transported down the axon (along microtubules) and that damaged mitochondria are transported back towards the cell body for degradation [52]. It will be interesting to test in future work how much of that scenario pertains to IMF in adult cardiac myocytes.
In conclusion, we have used live myocyte imaging to test for quantitative morphological and functional differences between IFM and PMN. We found that compared to IFM, PNM have lower rates of Ca2+ uptake, are less sensitive to oxidative stress, are smaller and rounder, and exhibit much more active translocation, fission and fusion. The relatively static IFM can however, be mobilized during stress (starvation or FCCP) to translocate via microtubular transport to the perinuclear region where mitophagy is more prominent. Thus, while IFM may be physiologically tuned to support local myofilament energy demands PNM may be more critical in mitochondrial turnover and regulation of nuclear-related functions. Thus, important functional differences are present in intrafibrillar vs. perinuclear mitochondrial subpopulations.
Methods
Cardiac Myocyte isolation, dye loading and permeabilization
Cardiac myocytes were isolated from New Zealand white rabbits and C57BL6 mice using retrograde Langendorff perfusion using Liberase TM (0.075 mg/mL, Roche) and Trypsin (0.0138%, Gibco) (37°C) as previously described [53]. All procedures were approved by the University of California Davis Institutional Animal Care and Use Committee (IACUC) in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Freshly isolated myocytes were plated on laminin-coated glass cover slip for >45 min before dye loading. All experiments were performed at room temperature (RT) (22–23°C). Cells were loaded with 2 μM Rhod-2 AM or 1 nM TMRM for 30 min, both at 23°C in nominally Ca free Tyrode’s solution (in mmol/L: HEPES 5, NaCl 140, KCl 140, MgCl2 1, glucose 10; pH adjusted to 7.4 with NaOH). 30 min were allowed for de-esterification. The ΔΨm in intact myocyte was measured by TMRM under field stimulation at 1 Hz with 1.8 mmol/L extracellular [Ca2+]. For plasma membrane permeabilization, cells were exposed to saponin (50 μg/ml; 30 sec) which was then washed off, in standard intracellular relaxing solution containing (in mmol/L) HEPES 10, K-aspartate 135, MgCl2 0.7, EGTA 2, reduced glutathione 10, MgATP 5, glucose 10, pH 7.2.
Viral transfection
Freshly isolated cells were plated on laminin-coated glass cover slips in serum-free PC-1 medium (Lonza) supplemented with penicillin/ streptomycin for 45 min before transfection. Mycoytes were exposed to adenoviral-mediated gene transfer of Mitycam, mtGFP, mtDsRed and mtPA-GFP for 4 hours at a multiplicity of infection (MOI) of 500 virus particles per cell (vp/cell), followed by replacement with fresh PC-1 media. Infected cells were cultured for 15 or 36 hr, with 1 final replacement of fresh medium 1 hr before experiments.
Transmission electron microscopy
Rabbit hearts were fixed by perfusion 2.5% (vol/vol) glutaraldehyde in 0.1 M Na-cacodylate (pH 7.4), the TEM image acquisition and analysis were performed as previous study [54].
Solutions
A Ca-buffered, Ca-free, Na-free internal solution contained (in mmol/L): EGTA 5, HEPES 20, K-aspartate 100, KCl 40, MgCl2 1, maleic acid 2, glutamic acid 2, pyruvic acid 5, KH2PO4 0.5, pH 7.2 adjusted with Trisma base. To control [Ca2+]i, 0.1 M CaCl2 solution was added as calculated with MaxChelator. Free [Ca2+] was confirmed by Ca-sensitive electrode. Na+-free internal solution with low Ca2+ buffering capacity and 100 nM free [Ca2+] contained (in mmol/L): EGTA 0.05, CaCl2 0.0234, HEPES 20, K-aspartate 100, KCl 40, MgCl2 0.551, maleic acid 2, glutamic acid 2, pyruvic acid 5, KH2PO4 0.5, MgATP 5, pH 7.2 adjusted with Trisma base. When NaCl was added to these solutions, equal amounts of KCl were omitted. 40 μM cytochalasin D was included to inhibit myocyte contraction. Starvation media (in mM): 20 HEPES, 110 NaCl, 4.7 KCl, 1.2 KH2PO4, 1.25 MgSO4, 1.2 CaCl2, 25 NaHCO3 [29].
Confocal imaging scanning Mitochondrial
Rhod-2, TMRM and mtDsRed fluorescence were measured in (Nikon A1 confocal microscope, 60× water-immersive objective, 535 nm excitation, emission at 560 ± 15 nm) The mtGFP, LysoTracker green signals were measured with same confocal (488 nm excitation, emission at 530 ± 15 nm). mtPAGFP signals were activated at 405 nm and fluorescence collected using excitation at 488 nm and emission at 530 ± 15 nm. Time-lapse x,y images were acquired at 512 bit resolution and at the sampling rate of 507 ms per frame.
Chemicals and statistics
Indicators and NBD C6-Ceramide were obtained from Invitrogen (Eugene, OR), LysoTracker green, LysoTracker red, MitoTracker green, DRAQ5 were obtained from Thermo Scientific (Waltham, MA) oligmycin and FCCP from Calbiochem (La Jolla, CA). To detect subcelluar distribution of fusion/fission proteins, anti-OPA1, Mfn1, Mfn2, Drp1 antibodies (Abcam) were used at 1:500 dilution. The secondary antibody carried fluorescein isothiocyanate derivative (Alexa Flour 488; Molecular Probes) and was used at 1:1000 dilution. Data are presented as mean ± SE of n measurements. Comparison between groups used Student’s t-test, One-Way ANOVA (significant at p<0.05).
Supplementary Material
Supplemental Figure 1 Morphological parameters for PN and IMF mitochondria in rabbit cardiac myocytes. Multiple shape descriptors were determined for perinuclear (PN) and intermyofibrillar (IF) mitochondria. n = 151 PN and 140 IF mitochondria.
Supplemental Figure 2 Subcellular distribution of mitochondrial fusion and fission proteins. Cardiac myoctyes were co-stained with MitoTracker green and Drp1(A), Mfn1 (B), Mfn2 (C) and OPA1 (D) antibodies (n=5-6 cells).
Highlights.
Within adult cardiac myocytes, mitochondria are morphological and functional heterogeneous.
Compared to perinuclear, intrafibrillar mitochondria exhibit higher [Ca]Mito uptake and more sensitive to oxidative stress.
Perinuclear mitochondria exhibit higher mobility, fission/fusion events and are critical in mitochondrial turnover.
Acknowledgments
Sources of Funding
The study was supported by National Institutes of Health grants R01-HL132831 and R01-HL030077 and National Natural Science Foundation of China grants 81741050 and 8187350.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Skulachev VP, Mitochondrial filaments and clusters as intracellular power-transmitting cables, Trends Biochemical Sci 26(1) (2001) 23–9. [DOI] [PubMed] [Google Scholar]
- [2].Anmann T, Eimre M, Kuznetsov AV, Andrienko T, Kaambre T, Sikk P, Seppet E, Tiivel T, Vendelin M, Seppet E, Saks VA, Calcium-induced contraction of sarcomeres changes the regulation of mitochondrial respiration in permeabilized cardiac cells, FEBS J 272(12) (2005) 3145–61. [DOI] [PubMed] [Google Scholar]
- [3].Hajnoczky G, Csordas G, Madesh M, Pacher P, The machinery of local Ca2+ signalling between sarco-endoplasmic reticulum and mitochondria, J Physiol 529 Pt 1 (2000) 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Garcia-Perez C, Schneider TG, Hajnoczky G, Csordas G, Alignment of sarcoplasmic reticulum-mitochondrial junctions with mitochondrial contact points, Am J Physiol Heart Circ Physiol 301(5) (2011) H1907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lu X, Ginsburg KS, Kettlewell S, Bossuyt J, Smith GL, Bers DM, Measuring local gradients of intramitochondrial [Ca2+] in cardiac myocytes during sarcoplasmic reticulum Ca2+ release, Circ Res 112(3) (2013) 424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Romashko DN, Marban E, O’Rourke B, Subcellular metabolic transients and mitochondrial redox waves in heart cells, Proc Natl Acad Sci (USA) 95(4) (1998) 1618–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhou L, Aon MA, Almas T, Cortassa S, Winslow RL, O’Rourke B, A reaction-diffusion model of ROS-induced ROS release in a mitochondrial network, PLoS Comput Biol 6(1) (2010) e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Palmer JW, Tandler B, Hoppel CL, Biochemical differences between subsarcolemmal and interfibrillar mitochondria from rat cardiac muscle: effects of procedural manipulations, Archiv Biochem Biophys 236(2) (1985) 691–702. [DOI] [PubMed] [Google Scholar]
- [9].Kasumov T, Dabkowski ER, Shekar KC, Li L, Ribeiro RF Jr., Walsh K, Previs SF, Sadygov RG, Willard B, Stanley WC, Assessment of cardiac proteome dynamics with heavy water: slower protein synthesis rates in interfibrillar than subsarcolemmal mitochondria, Am J Physiol Heart Circ Physiol 304(9) (2013) H1201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Palmer JW, Tandler B, Hoppel CL, Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle, J Biol Chem 252(23) (1977) 8731–9. [PubMed] [Google Scholar]
- [11].Riva A, Tandler B, Loffredo F, Vazquez E, Hoppel C, Structural differences in two biochemically defined populations of cardiac mitochondria, Am J Physiol Heart Circ Physiol 289(2) (2005) H868–72. [DOI] [PubMed] [Google Scholar]
- [12].Weinstein ES, Benson DW, Ratcliffe DJ, Maksem J, Fry DE, Experimental myocardial ischemia. Differential injury of mitochondrial subpopulations, Archiv Surgery 120(3) (1985) 332–8. [DOI] [PubMed] [Google Scholar]
- [13].Bowser DN, Minamikawa T, Nagley P, Williams DA, Role of mitochondria in calcium regulation of spontaneously contracting cardiac muscle cells, Biophys J 75(4) (1998) 2004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kuznetsov AV, Usson Y, Leverve X, Margreiter R, Subcellular heterogeneity of mitochondrial function and dysfunction: evidence obtained by confocal imaging, Molec cell Biochem 256–257(1–2) (2004) 359–65. [DOI] [PubMed] [Google Scholar]
- [15].Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ, The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis, Develop Cell 1(4) (2001) 515–25. [DOI] [PubMed] [Google Scholar]
- [16].Youle RJ, Karbowski M, Mitochondrial fission in apoptosis, Nat Rev Mol Cell Biol 6(8) (2005) 657–63. [DOI] [PubMed] [Google Scholar]
- [17].Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS, Fission and selective fusion govern mitochondrial segregation and elimination by autophagy, EMBO J 27(2) (2008) 433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee S, Jeong SY, Lim WC, Kim S, Park YY, Sun X, Youle RJ, Cho H, Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence, J Biol Chem 282(31) (2007) 22977–83. [DOI] [PubMed] [Google Scholar]
- [19].Huang X, Sun L, Ji S, Zhao T, Zhang W, Xu J, Zhang J, Wang Y, Wang X, Franzini-Armstrong C, Zheng M, Cheng H, Kissing and nanotunneling mediate intermitochondrial communication in the heart, Proc Natl Acad Sci (USA) 110(8) (2013) 2846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Eisner V, Cupo RR, Gao E, Csordas G, Slovinsky WS, Paillard M, Cheng L, Ibetti J, Chen SR, Chuprun JK, Hoek JB, Koch WJ, Hajnoczky G, Mitochondrial fusion dynamics is robust in the heart and depends on calcium oscillations and contractile activity, Proc Natl Acad Sci (USA) 114(5) (2017) E859–E868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu X, Weaver D, Shirihai O, Hajnoczky G, Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics, EMBO J 28(20) (2009) 3074–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sommer JR, Johnson EA, Ultrastructure of Cardiac Muscle, in: Berne RM (Ed.), Handbook of Physiology, Section 2, The Cardiovascular System, Amer. Physiol. Soc.1979, pp. 113–186. [Google Scholar]
- [23].Bers DM, Calcium cycling and signaling in cardiac myocytes, Ann Rev Physiol 70 (2008) 23–49. [DOI] [PubMed] [Google Scholar]
- [24].Territo PR, French SA, Dunleavy MC, Evans FJ, Balaban RS, Calcium activation of heart mitochondrial oxidative phosphorylation: rapid kinetics of mVO2, NADH, AND light scattering, J Biol Chem 276(4) (2001) 2586–99. [DOI] [PubMed] [Google Scholar]
- [25].Territo PR, Mootha VK, French SA, Balaban RS, Ca(2+) activation of heart mitochondrial oxidative phosphorylation: role of the F(0)/F(1)-ATPase, Am J Physiol. Cell Physiol 278(2) (2000) C423–35. [DOI] [PubMed] [Google Scholar]
- [26].Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ, Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury, Circulation 121(18) (2010) 2012–22. [DOI] [PubMed] [Google Scholar]
- [27].Wang J, Li J, Santana-Santos L, Shuda M, Sobol RW, Van Houten B, Qian W, A novel strategy for targeted killing of tumor cells: Induction of multipolar acentrosomal mitotic spindles with a quinazolinone derivative mdivi-1, Molec Oncol 9(2) (2015) 488–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bordt EA, Clerc P, Roelofs BA, Saladino AJ, Tretter L, Adam-Vizi V, Cherok E, Khalil A, Yadava N, Ge SX, Francis TC, Kennedy NW, Picton LK, Kumar T, Uppuluri S, Miller AM, Itoh K, Karbowski M, Sesaki H, Hill RB, Polster BM, The Putative Drp1 Inhibitor mdivi-1 Is a Reversible Mitochondrial Complex I Inhibitor that Modulates Reactive Oxygen Species, Develop Cell 40(6) (2017) 583–594 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA, Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells, Autophagy 6(4) (2010) 462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hoppel CL, Tandler B, Fujioka H, Riva A, Dynamic organization of mitochondria in human heart and in myocardial disease, Int J Biochem Cell Biol 41(10) (2009) 1949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liesa M, Palacin M, Zorzano A, Mitochondrial dynamics in mammalian health and disease, Physiological reviews 89(3) (2009) 799–845. [DOI] [PubMed] [Google Scholar]
- [32].Shimada T, Horita K, Murakami M, Ogura R, Morphological studies of different mitochondrial populations in monkey myocardial cells, Cell Tissue Res 238(3) (1984) 577–82. [DOI] [PubMed] [Google Scholar]
- [33].Hashitani H, Lang RJ, Suzuki H, Role of perinuclear mitochondria in the spatiotemporal dynamics of spontaneous Ca2+ waves in interstitial cells of Cajal-like cells of the rabbit urethra, Br J Pharmacol 161(3) (2010) 680–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hohendanner F, Maxwell JT, Blatter LA, Cytosolic and nuclear calcium signaling in atrial myocytes: IP3-mediated calcium release and the role of mitochondria, Channels (Austin) 9(3) (2015) 129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dalen H, An ultrastructural study of the hypertrophied human papillary muscle cell with special emphasis on specific staining patterns, mitochondrial projections and association between mitochondria and SR, Virchows Archiv. A, Pathol Anat Histopathol 414(3) (1989) 187–98. [DOI] [PubMed] [Google Scholar]
- [36].Kuznetsov AV, Margreiter R, Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity, Intl J Molec Scie 10(4) (2009) 1911–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN, Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription, Science signal 5(231) (2012) ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Glancy B, Hartnell LM, Combs CA, Femnou A, Sun J, Murphy E, Subramaniam S, Balaban RS, Power Grid Protection of the Muscle Mitochondrial Reticulum, Cell Rep 19(3) (2017) 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Westermann B, Bioenergetic role of mitochondrial fusion and fission, Biochim Biophys Acta 1817(10) (2012) 1833–8. [DOI] [PubMed] [Google Scholar]
- [40].O’Rourke B, Ramza BM, Marban E, Oscillations of membrane current and excitability driven by metabolic oscillations in heart cells, Science 265(5174) (1994) 962–6. [DOI] [PubMed] [Google Scholar]
- [41].Aon MA, Cortassa S, Marban E, O’Rourke B, Synchronized whole cell oscillations in mitochondrial metabolism triggered by a local release of reactive oxygen species in cardiac myocytes, J Biol Chem 278(45) (2003) 44735–44. [DOI] [PubMed] [Google Scholar]
- [42].Aon MA, Cortassa S, O’Rourke B, The fundamental organization of cardiac mitochondria as a network of coupled oscillators, Biophys J 91(11) (2006) 4317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kurz FT, Aon MA, O’Rourke B, Armoundas AA, Assessing Spatiotemporal and Functional Organization of Mitochondrial Networks, Methods Mol Biol 1782 (2018) 383–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhou B, Yu P, Lin MY, Sun T, Chen Y, Sheng ZH, Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits, J Cell Biol 214(1) (2016) 103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Robison P, Caporizzo MA, Ahmadzadeh H, Bogush AI, Chen CY, Margulies KB, Shenoy VB, Prosser BL, Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes, Science 352(6284) (2016) aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Drum BM, Yuan C, Li L, Liu Q, Wordeman L, Santana LF, Oxidative stress decreases microtubule growth and stability in ventricular myocytes, J Mol Cell Cardiol 93 (2016) 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Menzies RA, Gold PH, The turnover of mitochondria in a variety of tissues of young adult and aged rats, J Biol Chem 246(8) (1971) 2425–9. [PubMed] [Google Scholar]
- [48].Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, Zhang J, Zong NC, Lam MP, Ping P, Metabolic labeling reveals proteome dynamics of mouse mitochondria, Molec Cell Proteomics : MCP 11(12) (2012) 1586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Miwa S, Lawless C, von Zglinicki T, Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model, Aging Cell 7(6) (2008) 920–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Moyzis A, Gustafsson AB, Multiple recycling routes: Canonical vs. non-canonical mitophagy in the heart, Biochim Biophys Acta Mol Basis Dis (2018). [DOI] [PubMed] [Google Scholar]
- [51].Saito T, Nah J, Oka SI, Mukai R, Monden Y, Maejima Y, Ikeda Y, Sciarretta S, Liu T, Li H, Baljinnyam E, Fraidenraich D, Fritzky L, Zhai P, Ichinose S, Isobe M, Hsu CP, Kundu M, Sadoshima J, An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia, J Clin Invest (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Itoh K, Nakamura K, Iijima M, Sesaki H, Mitochondrial dynamics in neurodegeneration, Trends Cell Biol 23(2) (2013) 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Uchinoumi H, Yang Y, Oda T, Li N, Alsina KM, Puglisi JL, Chen-Izu Y, Cornea RL, Wehrens XHT, Bers DM, CaMKII-dependent phosphorylation of RyR2 promotes targetable pathological RyR2 conformational shift, J Mol Cell Cardiol 98 (2016) 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Picard M, White K, Turnbull DM, Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study, J Appl Physiol 114(2) (2013) 161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Morphological parameters for PN and IMF mitochondria in rabbit cardiac myocytes. Multiple shape descriptors were determined for perinuclear (PN) and intermyofibrillar (IF) mitochondria. n = 151 PN and 140 IF mitochondria.
Supplemental Figure 2 Subcellular distribution of mitochondrial fusion and fission proteins. Cardiac myoctyes were co-stained with MitoTracker green and Drp1(A), Mfn1 (B), Mfn2 (C) and OPA1 (D) antibodies (n=5-6 cells).