Fig. 2.
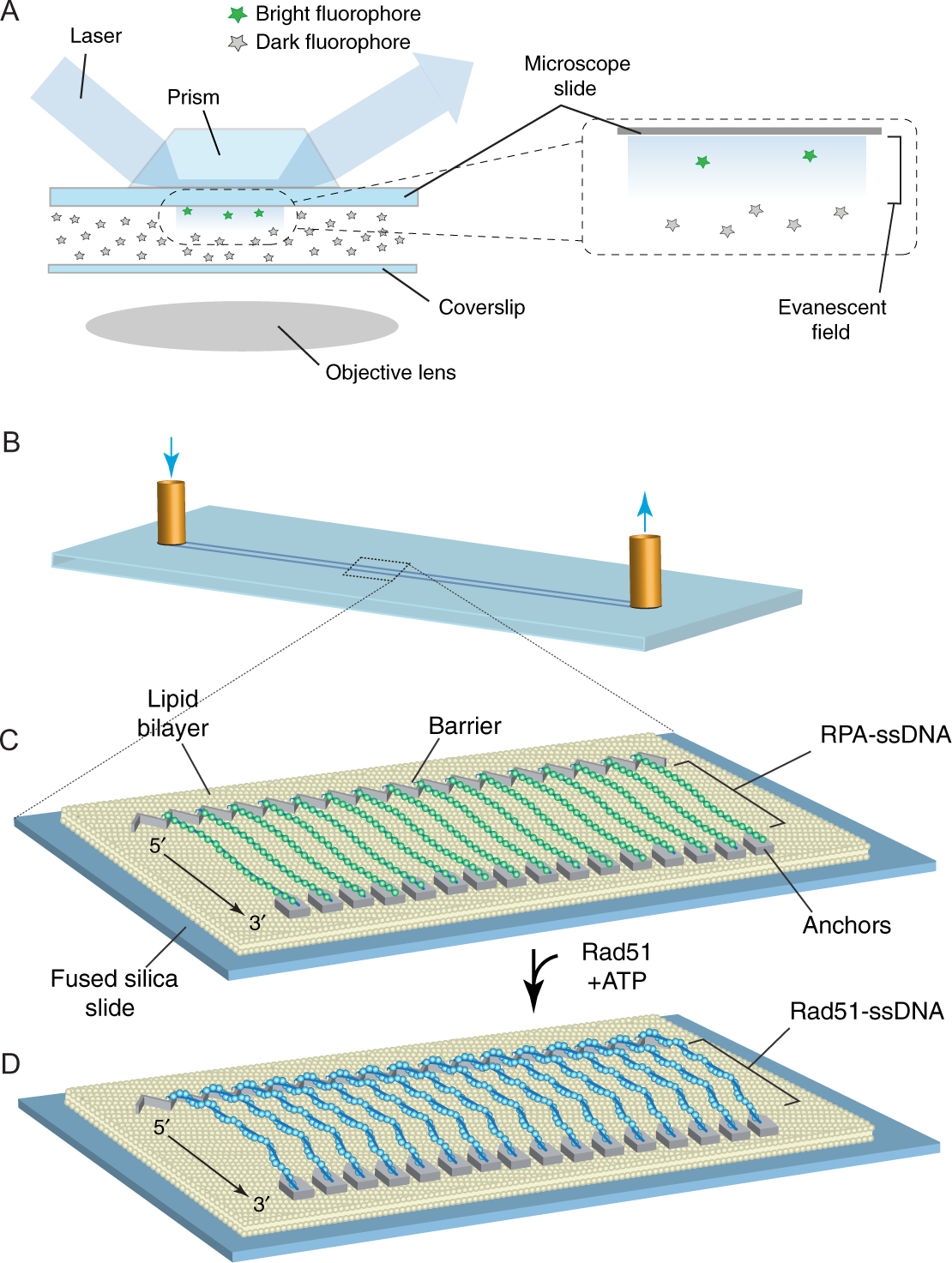
Schematic overviews of TIRFM and ssDNA curtains. (A) This illustration shows a simplified depiction of a typical prism-type TIRFM (Axelrod, 1989). Note: We use prism-type TIRFM setups because they allow for observation of reactions anchored to the microscope slide glass, as opposed to the coverslip, which is visualized in objective-type TIRFM. The benefit of prism-type TIRFM is that the thicker microscope slides are more robust and can be easily modified by electron-beam lithography, and then repeated cleaned and reused (Greene et al., 2010; Ma et al., 2017). (B) Schematic illustration of a flow cell used for sample analysis by TIRFM. (C) Schematic showing a double-tethered ssDNA curtain. In this example, the surface is coated with a lipid bilayer, which is disrupted at defined locations by metallic patterns that have been deposited by electron-beam lithography (Greene et al., 2010; Ma et al., 2017). The zigzag-shaped upstream pattern is used for alignment of the ssDNA, and the pentagonal anchor points allow for nonspecific adsorption of the downstream ends of the RPA-ssDNA molecules. In this configuration, the RPA-ssDNA can be viewed even in the absence of buffer flow. Similar patterns, but omitting the anchor points, can be used to set up single-tethered ssDNA curtains, in which case constant buffer flow is necessary to keep the molecules within the evanescent field for visualization. (D) Rad51-ssDNA filaments can be assembled by injecting Rad51 into the sample chamber in the presence of ATP, and assembly of these filaments coincides with the loss of GFP signal due to dissociation of the GFP-tagged RPA (Qi et al., 2015). Panels (B)–(D) were reproduced with permission from Qi, Z., Redding, S., Lee, J. Y., Gibb, B., Kwon, Y., Niu, H., et al. (2015). DNA sequence alignment by microhomology sampling during homologous recombination. Cell, 160, 856–869. doi: https://doi.org/10.1016/j.cell.2015.01.029.
