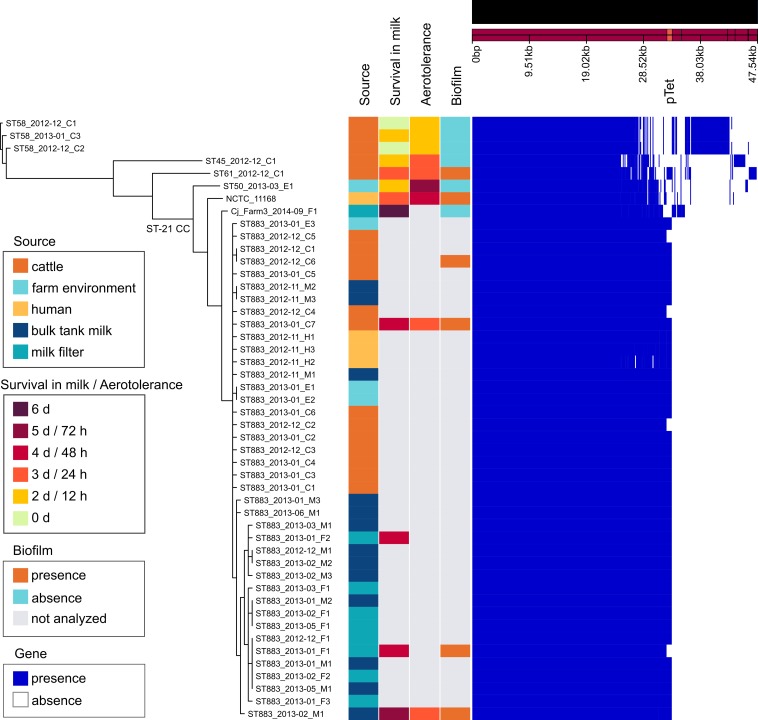
Fig 5. Genomic comparison of C. jejuni isolates from the outbreak farm.
The reference strain NCTC 11168 (RefSeq accession no. NC_002163.1) and ST-883 isolate (Cj_Farm3_2014–09_F1) from another dairy farm [11] are included in the comparison. (A) Left pane: Approximation of maximum-likelihood phylogeny based on the nucleotide alignment of 1356 core genes from Roary using FastTree (version 2.1.9) with the GTR+CAT model [13]. Branches with support <1 are not shown and recombinations are not masked. Names of the farm isolates indicate: ST, sampling time (year-month), sample source (C, E, H, M, or F; see legend), and isolate number. Middle pane: sample source and phenotype. Right pane: presence and absence of genes according to Roary.