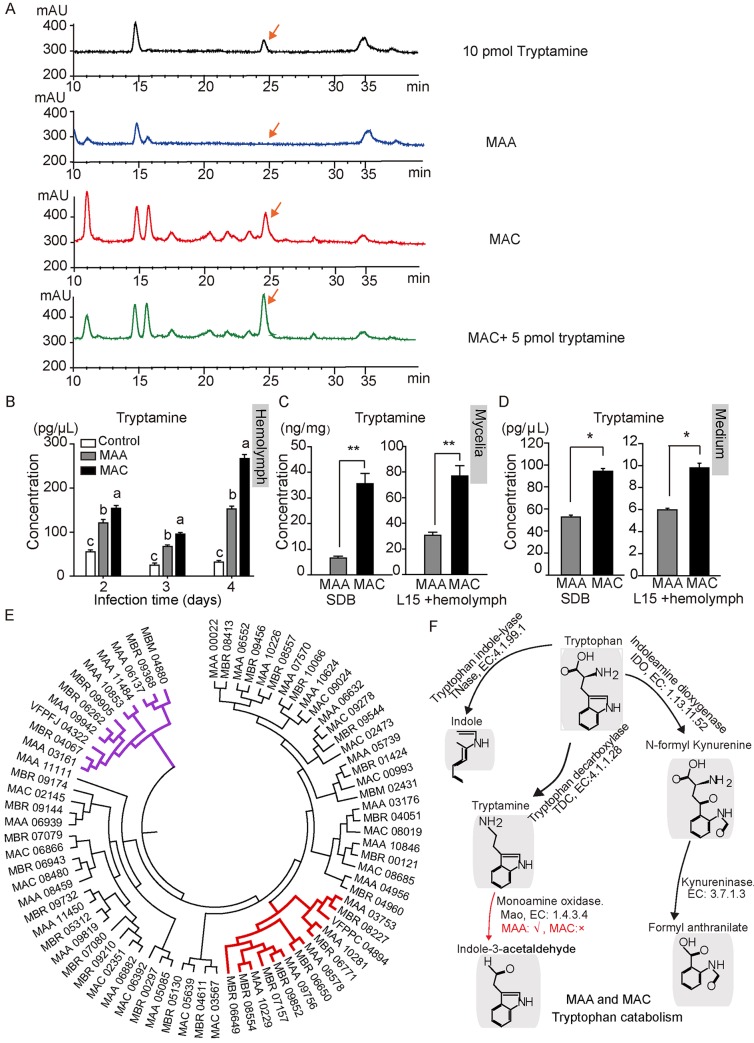
Fig 4. MAC produced a higher level of tryptamine than did MAA because of the absence of Mao genes.
(A) HPLC analysis of tryptamine level in mycelia of MAA and MAC. The results showed that the mycelia of MAA presented little of tryptamine while samples of MAC presented a high level of tryptamine. The tryptamine peak appeared at about 24.5 min. (B) HPLC analysis of the tryptamine level in the hemolymph of locusts after being infected by MAA and MAC at different times. The results showed that the tryptamine level increased according to the time of infection, and MAC infection induced the highest level of tryptamine in locusts (n = 12, different lowercase letters indicate the significance, P < 0.05, one-way ANOVA for analysis). (C) After being cultured in SDB medium for 3 days or in L-15 medium with locust hemolymph for 6 days, the MAC mycelia showed a higher level of tryptamine than those of MAA. nSDB = 7, nL15 = 18. Asterisks show the significance at P < 0.01. (D) HPLC analysis showed that the medium of MAC presented a higher level of tryptamine than that of MAA. (n = 10, asterisk indicated significance at P < 0.05 level). (E) Phylogeny analysis of protein sequences was used to classify the distribution of MAO proteins in Metarhizium species. Red lines show the mammal-like MAO; purple lines represent the FAD-dependent oxidases. The VFPPC_04894 (amine oxidase, Pochonia chlamydosporia) and VFPFJ_04322 (FAD-dependent oxidoreductase, Purpureocillium lilacinum) were used to indicate amine oxidase classification. The phylogeny tree was constructed via the neighbor-joining method by using MEGA 7.0 software. MAA: Metarhizium robertsii, MAC: Metarhizium acridum, MBR: Metarhizium brunneum, and MBM: Marssonina brunnea. (F) After analyzing the tryptophan catabolic pathway of MAA and MAC in the KEGG database, we found that MAC lost the Mao gene for tryptamine oxidation (EC: 1.4.3.4) in its genome, whereas MAA possessed three Mao (MrMao) genes.