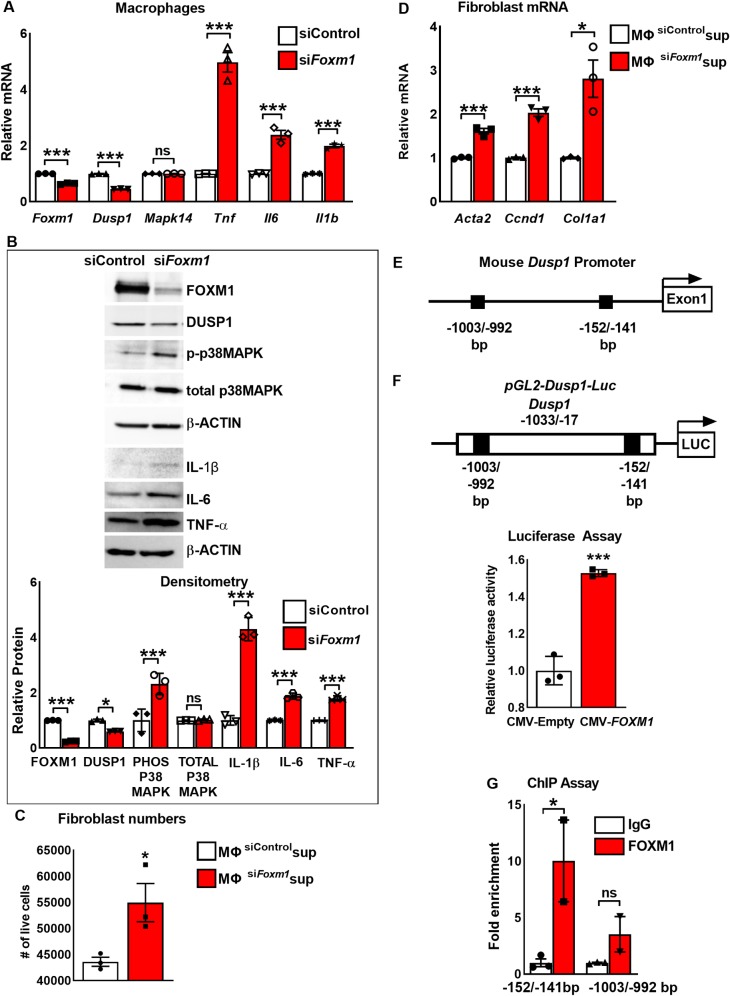
Fig 5. FOXM1 directly binds to and induces transcriptional activity of Dusp1 promoter in macrophages.
(A) Depletion of Foxm1 in macrophages decreased Dusp1 mRNA and increased Tnf, Il6 and Il1b mRNA. Mapk14 mRNA was unchanged. RAW264.7 macrophages were transfected with non-targeting siRNA (siControl) or siFoxm1. mRNA levels were analyzed by qRT-PCR. Actb mRNA was used for normalization (n = 3). (B) Depletion of Foxm1 from macrophages results in activation of p38 MAPK signaling pathway. RAW264.7 macrophages were transfected with non-targeting siRNA (siControl) or siFoxm1 and protein levels were analyzed by Western blot (top panel). FOXM1, DUSP1 protein levels were decreased and phospho-p38 MAPK, TNF-α, IL-1β and IL-6 levels were increased in Foxm1-deficient RAW264.7 macrophages. No changes were detected in total p38 MAPK. β-ACTIN was used as loading control. Blots are representative of three independent experiments. Densitometry was used to quantify Western blot results (bottom panel). Protein levels were normalized to β-ACTIN. Protein expression is presented as fold change over siControl transfected macrophages. (C) Supernatant from Foxm1-depleted RAW264.7 macrophages increases the number of viable 3T3 fibroblasts. 3T3 cells were cultured in supernatant obtained from RAW264.7 cells transfected with control siRNA or siFoxm1. 48 hours after transfection, the number of live cells was determined by trypan blue (n = 3). (D) Supernatant from Foxm1-depleted RAW264.7 macrophages increases Acta2, Ccnd1, and Col1a1 mRNA levels in 3T3 fibroblasts. mRNA levels were measured by qRT-PCR. Actb mRNA was used for normalization (n = 3). (E) Schematic representation of the mouse Dusp1 promoter region with potential FOXM1 DNA-binding sites (black boxes). (F) Schematic representation of the pGL2-Dusp1-Luc construct containing the Dusp1 promoter region. The mouse Dusp1 promoter (-1033/-17 bp) was cloned into the pGL2-LUC vector and co-transfected with CMV-Empty vector (CMV-EV) or CMV-FOXM1. Luciferase assay shows that FOXM1 transcriptionally activates the Dusp1 promoter. Data represent mean ± SEM of three independent experiments. (G) ChIP assays shows direct binding of FOXM1 to the -152/-141bp Dusp1 promoter region. FOXM1 binding is shown relative to immunoglobulin G (IgG). Data represent mean ± SEM of two independent experiments. * = P > 0.05; ** = P > 0.01, *** = P < 0.001, by Student’s t-test.