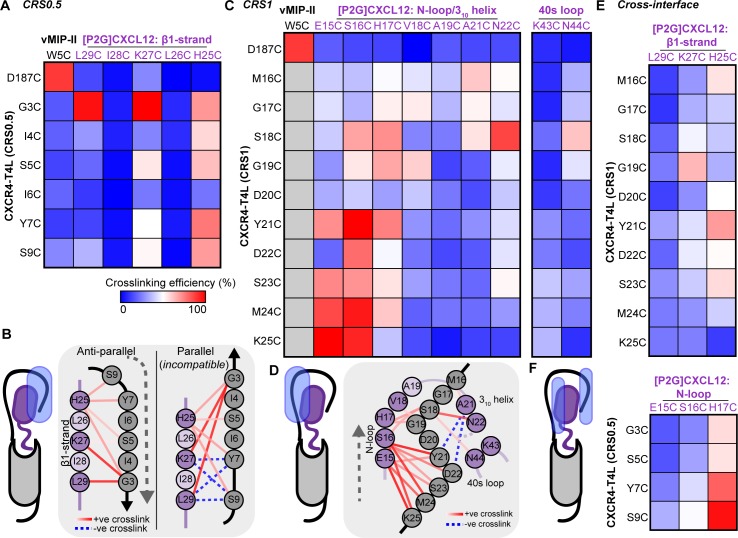
Fig 2. CXCR4-[P2G]CXCL12 intermolecular crosslinking efficiencies detected by flow cytometry.
(A) A heat map of pairwise cross-linking efficiencies in the predicted CRS0.5, normalized to CXCR4(D187C)-vMIP-II(W5C), a previously crystallized covalent complex. (B) CRS0.5 crosslinking efficiencies mapped onto a 2D schematic of CRS0.5 in the hypothetical antiparallel and parallel β-sheet orientations. The data are more consistent with the antiparallel geometry. (C) A heat map of pairwise cross-linking efficiencies in CRS1, normalized to CXCR4(D187C)-vMIP-II(W5C). (D) CRS1 crosslinking efficiencies mapped onto a 2D schematic of CRS1. (E–F) Heat maps of pairwise cross-linking efficiencies when (E) Cys mutants in CXCR4 CRS1 are co-expressed with Cys mutants in CXCL12 β1-strand (CRS0.5) and (F) vice versa. Gray squares in the heat maps represent residue pairs that were not studied. Data represent the average of n ≥ 3 independent biological replicates. The data in numerical form are provided in S4 Table, and the underlying numerical data for S4 Table and figure panels are in S1 Data. CRS, chemokine recognition site.