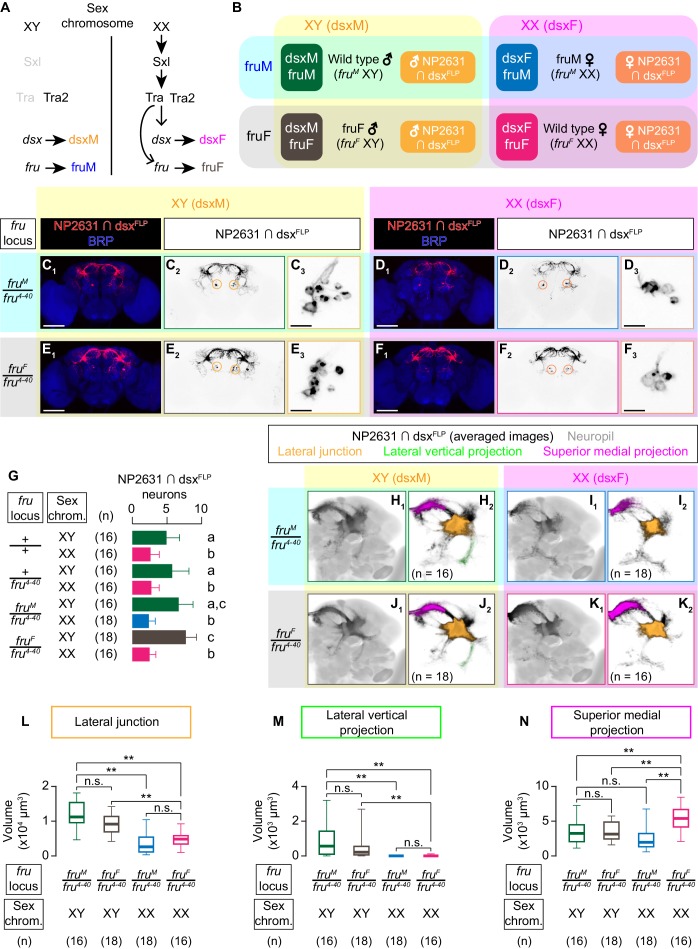
Figure 3. dsx specifies the sexual dimorphism of NP2631 ∩ dsxFLP neurons.
(A) Schematics of the sex-determination pathway in Drosophila. (B) Schematic of the four sex genotypes defined by dsx and fru splicing, and how NP2631 ∩ dsxFLP neurons are specified in each genotype (see following panels for details). (C–F) Expression of CsChrimson:tdTomato under the control of NP2631 and dsxFLP (red in C1–F1, black in C2,3-F2,3) in brains of a male (C), fruM female (D), fruF male (E), and female (F) is visualized together with a neuropil marker BRP (blue) by immunohistochemistry. Circle: soma (right cluster is enlarged in C3–F3). Scale bar: 100 μm (C1–F1), 10 μm (C3–F3). (G) Mean number of cell bodies per hemibrain visualized by anti-DsRed antibody in each genotype represented in C–F) and Figure 3—figure supplement 1A–D. (H–K) Z-projection of the registered and averaged images of CsChrimson:tdTomato expression under the control of NP2631 and dsxFLP (black in H1–K1) in male (H), fruM female (I), fruF male (J), and female (K). A part of the standard Drosophila brain is shown in gray in H2–K2). Number of used hemibrains are indicated in H2–K2). Lateral junction (orange), lateral vertical projection (green), and superior medial projection (magenta) are segmented and overlaid in H2-K2. L-M: Boxplot of volumes of lateral junction (L), lateral vertical projection (M), and superior medial projections (N). Genotypes and number of hemibrains are indicated below the plot. **p<0.01, n.s. p>0.05 (Kruskal-Wallis one-way ANOVA and post-hoc Wilcoxon signed rank test).