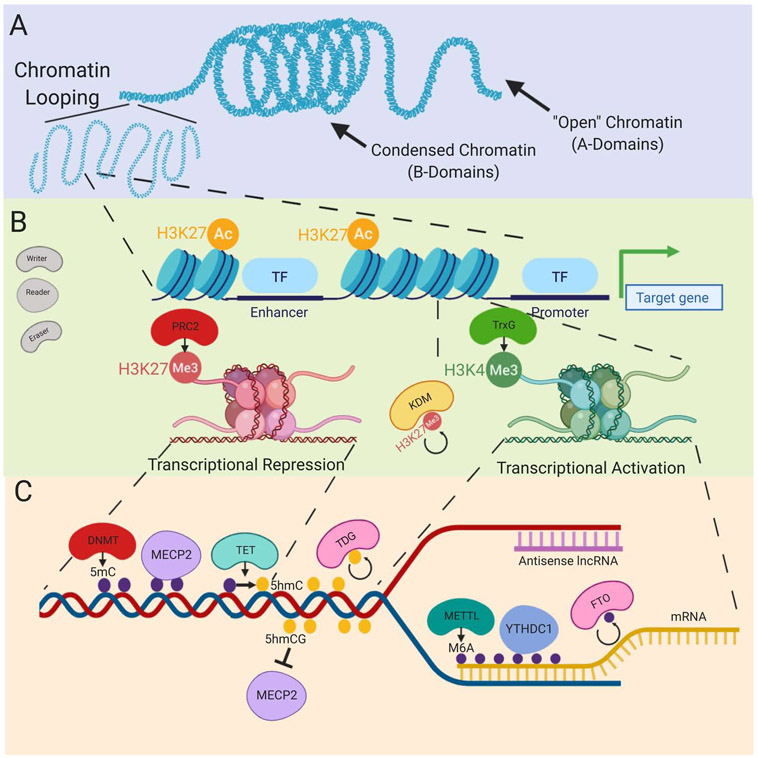
Figure 2. Epigenetic Mechanisms of Gene Regulation.
(A) Epigenetic regulation of higher ordered structures begins on the level of accessibility of chromatin. A-type chromatin (euchromatin) refers to open chromatin that is accessible for transcription, while B-type (heterochromatin) chromatin refers to closed chromatin that is typically association with gene inactivity. These areas of open chromatin can form chromatin loops that generate 3-dimensional structures that allow for long-range interactions that are not directly observable from the 2-dimensional sequence. (B) Long range interactions generated by 3-dimensional chromatin loops can form enhancer complexes, wherein transcription factors can bind to enhancer regions that are distal to the promoter region in 2-dimensions to influence gene expression. These enhancer regions are associated with histone modifications including H3K27ac (pictured), H3K4me1, as well as the enhancer-associated p300 protein. Promoter regions can be either inactive, which is associated with the Polycomb Repressive Complex-group catalysed H3K27me3 (pictured) or active, which is associated with the Trithorax group catalysed H3K4me3 (pictured). Regions with both H3K27me3 and H3K4me3 are considered inactive but poised for transcription. Removal of H3K27me3 by histone demethylases (KDM family proteins) can lead to transcriptional activation. (C) DNA methylation is written by DNA methyltransferases (DNMTs) and is traditionally associated with gene silencing due to interactions with MECP2. 5mC DNA methylation is removed through oxidation to 5hmC by TET family proteins and subsequent TDG-mediated excision, leading to base excision repair. MECP2 does not bind to 5hmCG, which is associated with regions of transcriptional activation. Transcription can be regulated by m6A modification on the RNA strand. This modification is written by METTL family proteins, read by various readers including YTHDC1, and enzymatically removed by various erases including FTO. m6A modification has diverse roles including regulating mRNA metabolism and translation and mediating RNA nuclear transport. Non-coding RNAs, including long-noncoding RNAs (lncRNAs), frequently occur on the anti-sense strand of mRNAs. These lncRNAs can operate locally in cis or distal to the area of transcription in trans in order to regulate gene expression.