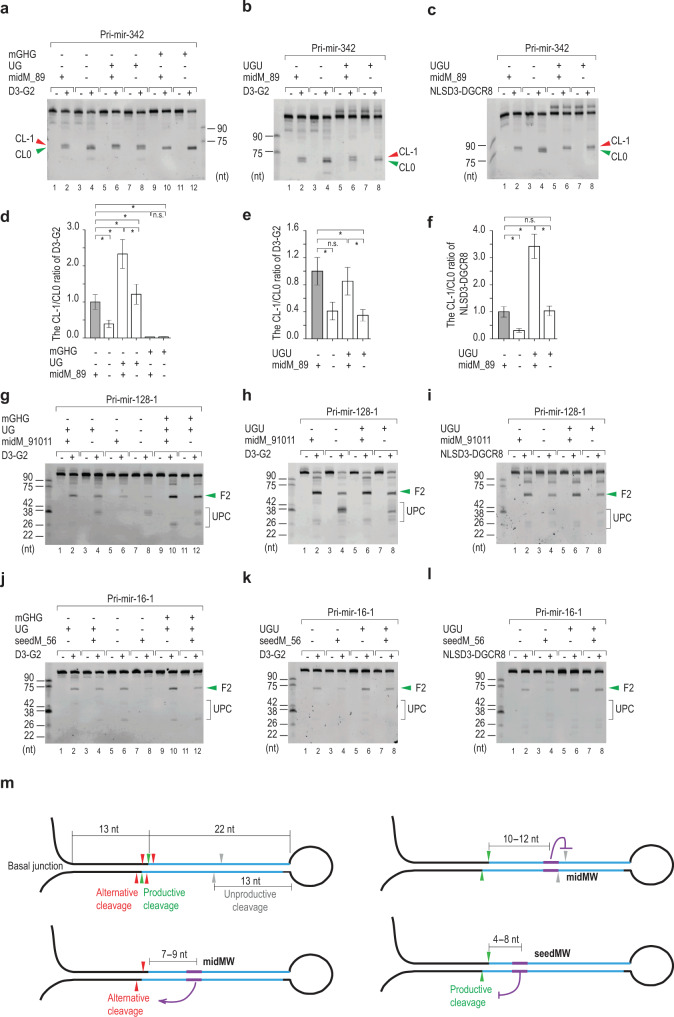
Fig. 7. Coordination of midMW and seedMW with UG, UGU, and mGHG in determining the accuracy and efficiency of Microprocessor cleavages.
a–c Processing of pri-mir-342 variants by D3-G2 or NLSD3-DGCR8. Each RNA (6 pmol) was incubated with D3-G2 (5 pmol) or NLSD3-DGCR8 (3 pmol) for 60 min at 37 °C in 10 μl standard reaction buffer. d–f Bar graphs to show the relative alternative cleavage activity, which was estimated from experiments conducted in triplicate as a ratio of CL-1 to CL0 cleavage. The band densities of the F2 fragments resulting from the CL-1, and CL0 cleavages were measured using Image Lab v.6.0.1. Data are presented as mean values +/− SEM. The asterisks (*) and n.s. indicate statistical significant differences and no statistical significant differences, respectively, from the two-sided t-test (d midM_none vs. midM_89: p = 0.002, midM_89_UG vs. midM_89: p = 1.1e-5, midM_none_UG vs. midM_89: p = 0.006, midM_89_GHG vs. midM_89: p = 0.0013, midM_none_GHG vs. midM_89: p = 0.046, midM_89_UG vs. midM_none_UG: p = 0.001, midM_89_GHG vs. midM_none_GHG: p = 0.923; e midM_none vs. midM_89: p = 0.009, midM_89_UGUG vs. midM_89: p = 0.064, midM_none_UGUG vs. midM_89: p = 0.011, midM_89_UGUG vs. midM_none_UGUG: p = 0.005; f midM_none vs. midM_89: p = 1.7e-4, midM89_UGUG vs. midM_89: p = 1.5e-5, midM_none_UGUG vs. midM_89: p = 0.472, midM_89_UGUG vs. midM_none_UGUG: p = 2.3e-4). g–l Processing of pri-mir-128-1 (g–i) and pri-mir-16-1 (j–l) variants by D3-G2 or NLSD3-DGCR8. Each RNA was added with D3-G2 (10 pmol) or NLSD3-DGCR8 (6 pmol). Pri-mir-128-1 and pri-mir-16-1 substrates were incubated at 37 °C in 10 μl standard reaction buffer for 120 and 30 min, respectively. m Mismatched and wobble base pairs in the upper stem of pri-miRNA govern the accuracy and efficiency of Microprocessor. The midMW that contains mismatches and/or wobble base pairs in the middle of the miRNA sequence stimulates the alternative cleavages and/or blocks the unproductive cleavages of Microprocessor when located at 7–9 nt and 10–12 nt in the upper stem, respectively. The seedMW that possesses mismatches and/or wobble base pairs in the seed region of the 5p miRNA sequence inhibits the productive cleavages of Microprocessor. The green, red, and gray arrowheads indicate the productive, alternative, and unproductive cleavages, respectively. The blue lines show the miRNA sequence embedded in pri-miRNA. UPC: unproductive cleavage. The source data are provided in the Source Data file.