Fig. 5. Distribution of YNP-TEG-like phylotypes among YNP hot spring communities.
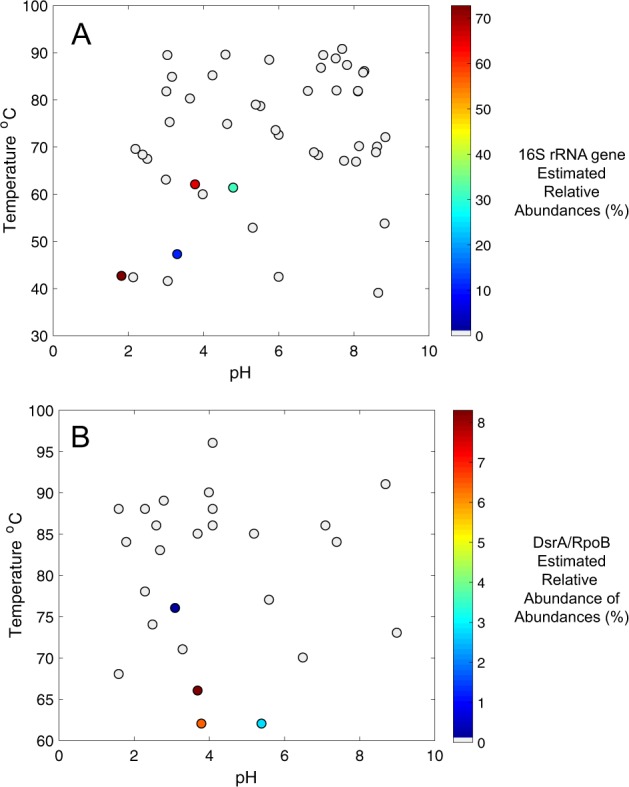
a 16S rRNA gene distribution revealed by comparison against 32 YNP hot spring communities described in Colman [26] and 15 communities evaluated in Colman et al. [25]. Positive detection was defined as when 16S rRNA gene identities >97% were identified relative to the MV2-Eury 16S rRNA gene. Points are colored by estimated relative abundance, as given by the scale on the right. Gray circles indicate the lack of detection in >0.01% relative abundance. b Inferred distribution of YNP-TEG-like phylotypes using an in-house database of 33 chemosynthetic hot spring community metagenomes that we have previously generated from YNP springs (Supplementary Table S4). Relative abundances were estimated based on DsrA/RpoB amino acid identities >95% to the MV2-Eury phylotype in MAG-binned contigs, followed by relative abundance estimates calculated by read-mapping as described in the materials and methods. The distribution of MV2-Eury-like DsrA and RpoB homologs were identical, and thus only one scatter plot is shown. Of the four metagenomes with positive YNP-TEG detection, two were MV2 from different years and another was SJ3 (Supplementary Table S4). Note that the samples in a and b derive from different datasets and do not necessarily represent the same springs. Likewise, the temperature and relative abundance scales differ between the two panels to emphasize within-dataset variation.
