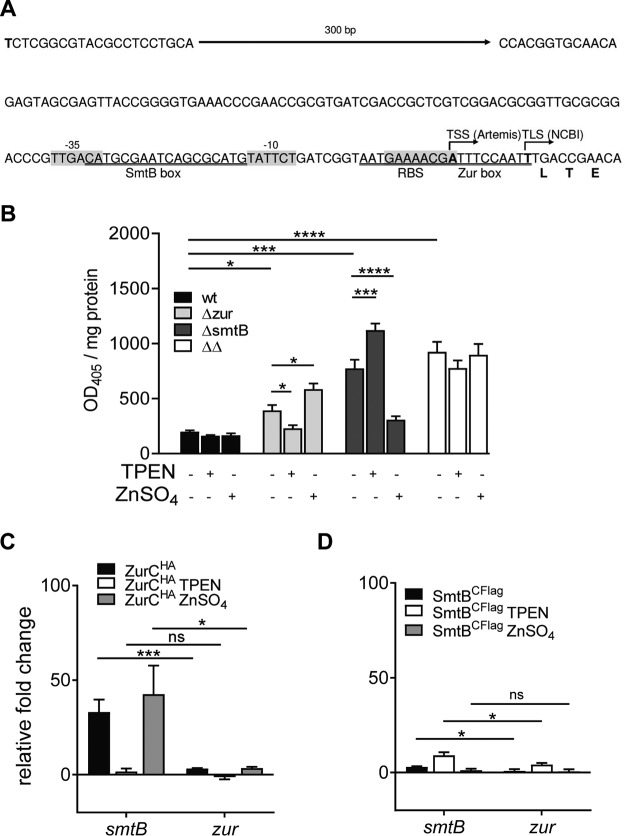
FIG 5.
Analysis of smtB-zur regulation. (A) Schematic depiction of the smtB-zur promoter region (NC_008596; positions 4568740 to 4569210). The putative transcription start site (TSS), determined by RNA-Seq and subsequent Artemis analysis, and the putative translation start site (TLS) according to NCBI are in bold. Putative −10 and −35 promoter sites, predicted by BDGP, as well as a ribosome binding site (RBS) are highlighted in gray. Zur box (underlined) prediction was accomplished using the MEME Suite and subsequent FIMO analysis. SmtB box (underlined) was predicted by Canneva et al. (30). (B) β-Galactosidase assay. MSMEGwt (black bars), MSMEGΔzur (light gray bars), MSMEGΔsmtB (dark gray bars), and MSMEGΔsmtBΔzur (white bars) were transformed with pJEM15-smtB1 harboring a 5′ UTR fragment as shown in panel A. Strains were grown in MB, incubated with 100 μM ZnSO4 or 10 μM TPEN for 24 h or left untreated, and lysed, and promoter activity was determined as described in the legend to Fig. 4. Shown are the results of three independent experiments (mean ± SEM) in triplicate after 21 min of incubation. Statistical analysis was performed by using one-way ANOVA with P values of <0.05 (*), <0.0005 (***), and <0.0001 (****). (C) ChIP assay. MSMEGΔzur was complemented with pMV306-Zur-HA and treated with 10 μM TPEN (white bars) or 500 μM ZnSO4 (gray bars) for 2 h or left untreated (black bars). ChIP from cross-linked lysates was performed using anti-HA and anti-IgG1κ (isotype control). DNA from ChIP and input control was subjected to qRT-PCR with primers targeting the smtB promoter (MSMEG_4486) or an intragenic region (zur, MSMEG_4487). (D) SmtB-ChIP assay was performed as described for Zur but with MSMEGΔsmtB complemented with pMVM306-SmtB-Flag and precipitated with anti-Flag and anti-IgG2a. Shown are the results of at least three independent experiments in duplicate. Statistical analysis was performed using Student's t test (unpaired) with P values of <0.05 (*), <0.0005 (***), and <0.0001 (****).