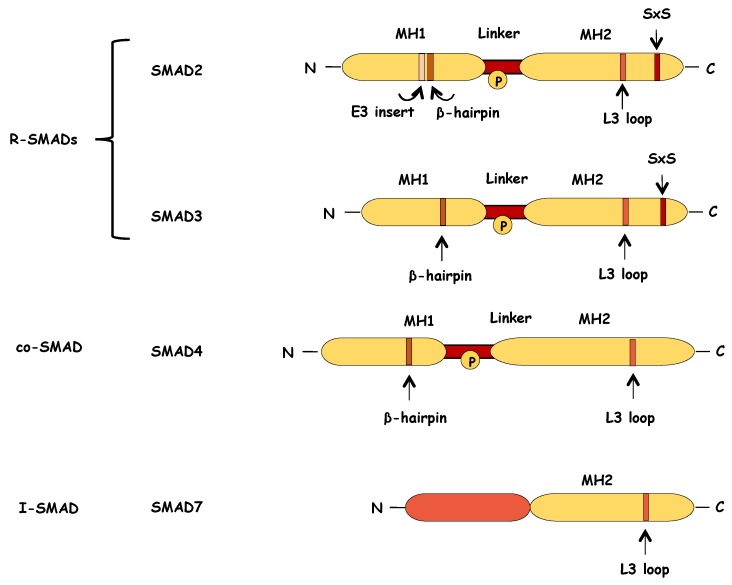
Figure 5.
Schematic representation of structure of the SMAD proteins. R-SMAD and SMAD4 proteins consist of two highly conserved domains, the MH1 and MH2 domains, which are separated by a non-conserved linker region. The N-terminal MH1 domain of R-SMADs contains a β-hairpin structure that is critical for DNA binding. In the case of SMAD2, the MH1 domain contains also an extra amino-acid sequence (E3 insert) that negatively regulates the DNA binding capacity of SMAD2. The C-terminal MH2 domain of R-SMADs contains an L3 loop that mediates the interaction between R-SMADs and the activated type I receptor. This L3 loop is also part of the SMAD4 structure and it is important for the formation of SMAD trimeric complexes. At their very C-terminus, R-SMADs, have a short conserved motif of two serines separated by one amino acid (Ser-X-Ser (SXS)) that are phosphorylated by the activated type I receptor, thus leading to the R-SMAD activation. The linker region encompasses multiple phosphorylation sites (P in circle), and it is targeted by various kinases that modulate SMAD stability and function. SMAD7, the inhibitory SMAD, retains the conserved MH2 domain but lacks the SXS motif at the C-terminus and the N-terminal region presents small similarity to the MH1 domain.