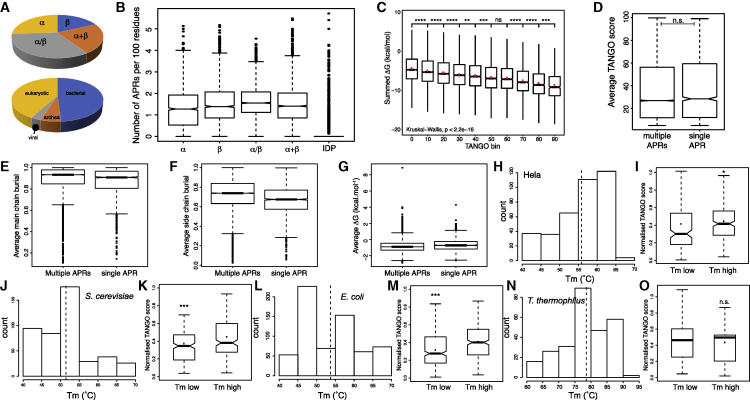
Figure 1.
Stability and Aggregation Propensity Are Related
(A) Class and kingdom composition of the SCOPe dataset.
(B) Boxplot representation of the distribution of APRs in the SCOPe and IDP datasets.
(C) Boxplot showing the contribution of APRs to the stability of the native state calculated by FoldX in the SCOPe dataset in function of the predicted aggregation propensity by TANGO.
(D–G) Boxplots comparing APRs occurring in domains with one APR to those occurring in domains with more than one APR: the distribution TANGO score of APRs (D), the average main-chain burial (E), the average side-chain burial (F), and the average contribution of an APR to native-state stability (G, ΔG calculated by FoldX, in kilocalories per mole).
(H, J, L, and N) Histograms of the melting temperature (Tm) observed in whole-proteome protein stability measurements (Leuenberger et al., 2017) for HeLa cells (H), S. cerevisiae (J), E. coli (L), and T. thermophilus (N). The dotted line indicates the mean Tm of the proteome in question.
(I, K, M, and O) Boxplots comparing the normalized TANGO scores of proteins with a high or low Tm value in HeLa cells (I), S. cerevisiae (K), E. coli (M), and T. thermophilus (O).
The bottoms and tops of the boxes are the first and third quartiles, and the band inside the box represents the median; the dot indicates the mean. The whiskers encompass the minimum and maximum of the data. Significant differences were computed using a Wilcox rank test. Asterisks denote level of significance: n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The source files (Data S1) and R-scripts (Data S2) used to generate this figure are available.