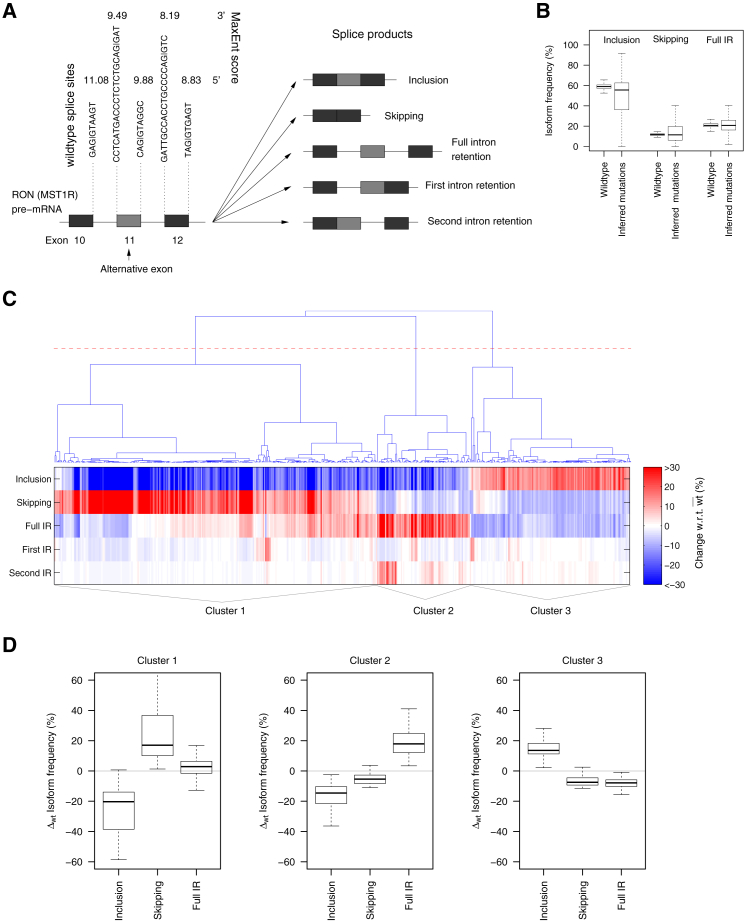
Figure 1.
Sequence mutations in a three-exon minigene containing RON AE 11 induce concerted changes in the distribution of splice isoforms. (A) The studied three-exon-minigene (704 bp) contains RON AE 11 and the complete adjacent introns and constitutive exons 10 and 12. The wild-type (wt) sequences of the 3′ and 5′ splice sites included in the minigene are shown together with the corresponding MaxEnt splice site scores. Using next-generation sequencing, five different splice products were quantified (as percent of all splice products) for wt minigenes as well as for single point mutations (see Methods and (19)). (B) Point mutations induce strong changes in the splice isoform distribution, as visible by the much broader isoform frequency distributions of the mutated minigenes compared to the population of 500 unmutated wt minigenes. Full IR: full intron retention. (C) A heat map of splice isoform difference between mutant and wt is plotted for 510 point mutations (columns) with a strong effect on the splicing (more than 10% change in at least one isoform frequency with respect to wt). Mutations are sorted using hierarchical clustering (cosine distance), and three main clusters are defined (using the red line in the dendrogram as a threshold). (D) The same data as in (C) are given, represented as boxplots summarizing the isoform distribution of each cluster for the three main isoforms. Mutations in clusters 1 and 3 induce anticorrelated changes in inclusion and skipping. In cluster 1, these changes are most pronounced in absolute terms, and IR is only slightly changed compared to wt. Cluster 3 shows weaker changes and altered IR, though in opposite direction. Mutations assigned to cluster 2 decrease both inclusion and skipping and simultaneously increase full IR.