Figure 2.
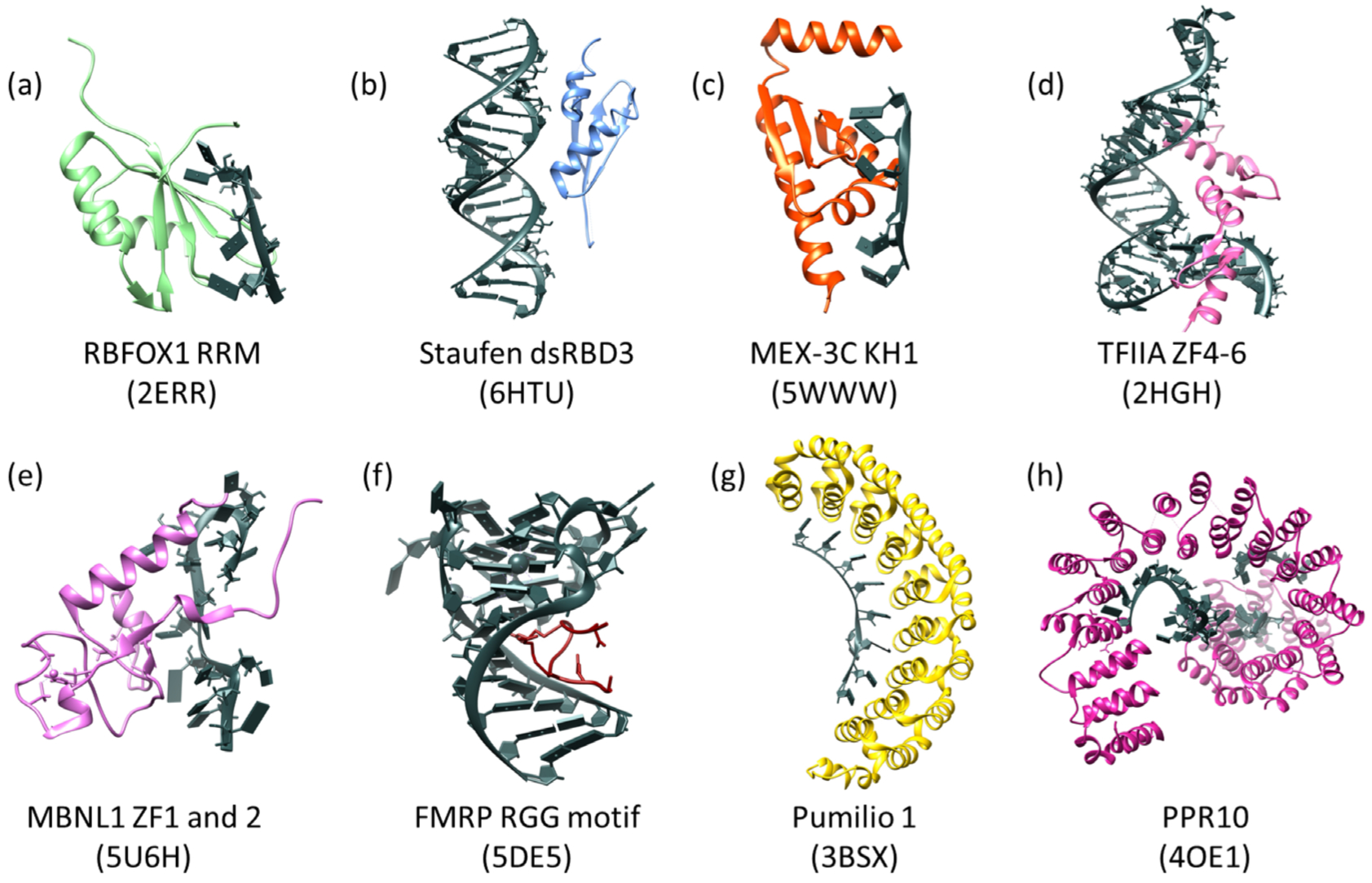
Example RBDs with RNA substrates (shown in dark gray with block nucleotides). (a) The RRM of human RBFOX1 in complex with a 7-mer oligo (UGCAUGU) which interacts through base stacking of aromatic residues and through ionic interactions. (b) DsRBD3 of human Staufen in complex with ARF1 RNA. Recognition through the specific shape of A-form dsRNA and the 2′OH present on the RNA. (c) The KH1 domain of human MEX-3C in complex with a 10-mer RNA oligo. Recognition through hydrogen bonding and shape complementarity. (d) Transcription Factor IIIA zinc fingers 4–6 from Xenopus laevis bound to 5S rRNA (55-mer). Recognition through base stacking of aromatic residues with RNA bases. (e) ZF1 and 2 of human MBNL1 in complex with RNA from Cardiac Troponin T, which interacts with the RNA via base stacking of aromatic residues and hydrogen bonding. (f) The human FMRP RGG motif in complex with G-quadruplex RNA of sc1, which interacts with the RNA via the arginines present in these domains. (g) Human Pumilio 1 in complex with Puf5 RNA. This domain interacts with RNA via base stacking and hydrogen bonds. (h) Zea mays PPR10 in complex with an 18-nt PSAJ RNA element. This domain interacts with RNA similar to PUF domains in that it utilizes hydrogen bonding and base stacking. The PDB entrys are below the names in parentheses for all domains. All panels were made using Chimera (Pettersen EF 2004).
