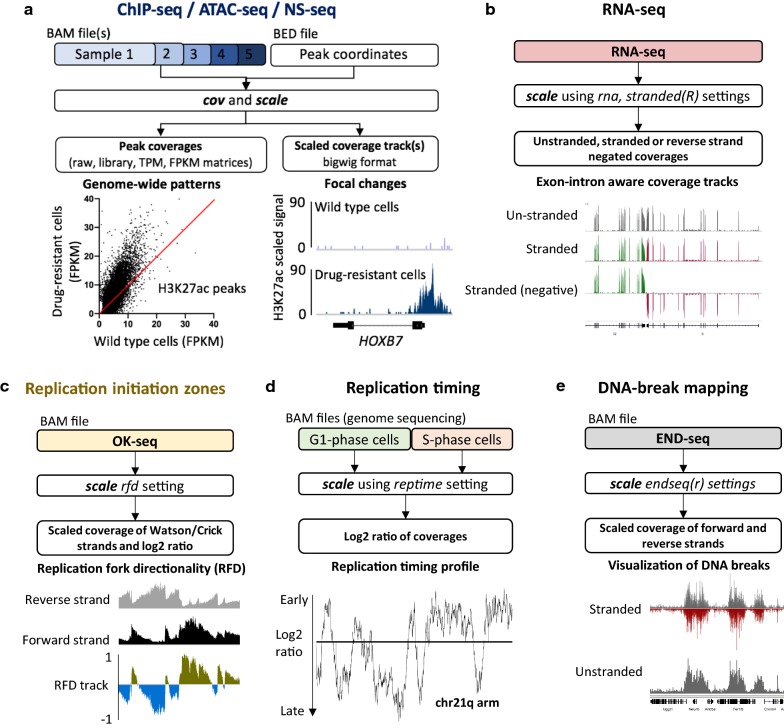
Fig. 1.
Application and benchmarking of BAMscale on different sequencing datasets. a Scaled coverage track generation and peak quantification of ChIP-seq and ATAC-seq data. Local differential H3K27ac signal at the HOXB7 locus in MV4-11 (wild type) and PKC412-resistant (R) (drug-resistant) cells, and global H3K27ac increase. b Creating exon–intron boundary aware stranded and un-stranded RNA-seq coverage tracks. c OK-seq coverage track creation using BAMscale outputs scaled strand-specific coverage tracks, and the replication-fork directionality tracks. d Analysis of replication timing data. e Mapping DNA-breaks from END-seq, creating strand-specific or unstranded coverage tracks