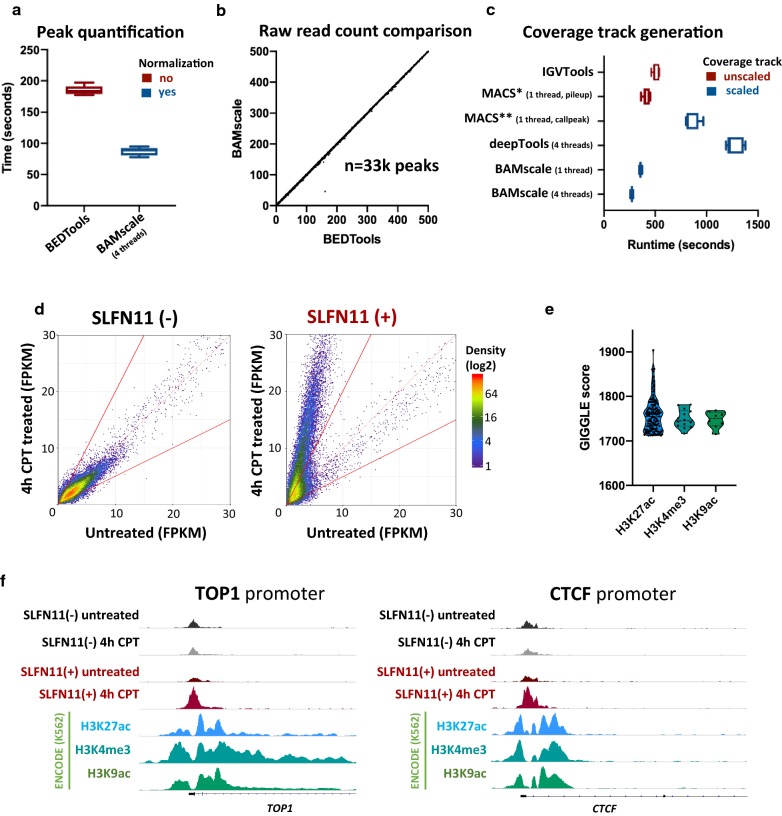
Fig. 2.
Benchmarking BAMscale on ATAC-seq data. a Performance comparison of peak quantification and b correlation of raw read counts in ~ 33 k peaks between BAMscale and bedtools. c Coverage tracks generation benchmarks using IGVtools (unscaled output), MACS2 (pileup: unscaled, callpeak: scaled with peak calling), deeptools and BAMscale (scaled output). d ATAC-seq signal change induced from camptothecin (CPT) is observed in wild-type CEM-CCRF cells (SLFN11 positive), and not in the SLFN11 isogenic knockout. e Colocalization of opening ATAC-seq peaks using GIGGLE and Cistrome. f Examples of chromatin accessibility in the TOP1 and CTCF genes