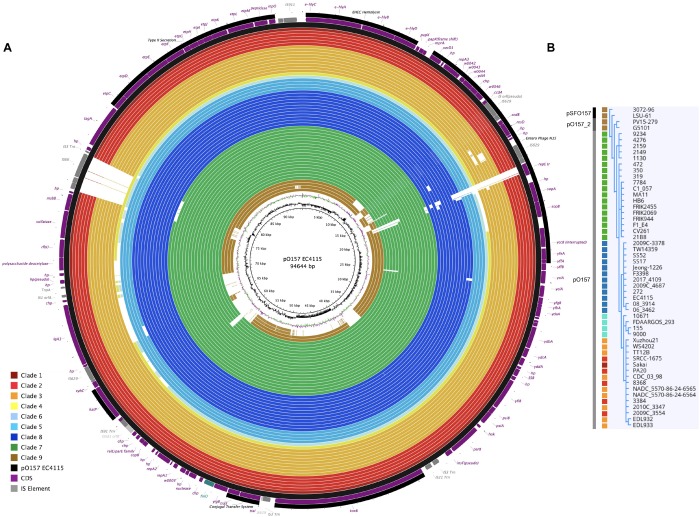
FIGURE 5.
Comparison of virulence plasmid variants referenced to pO157. (A) BRIG visualization (Alikhan et al., 2011) of plasmid architectures and gene inventories comparing the three major virulence plasmid variants. Only draft sequences were available for clades 4 and 6 plasmids, while closed molecules represent all other plasmids. Plasmids recovered and sequenced in this study were complemented by O157:H7 and SF O157:NM plasmid sequences retrieved from GenBank, and referenced to the 94-kb pO157 plasmid type of strain EC4115, shown on the outermost circle. CDS are presented as purple arrows and functional annotation for loci of importance are depicted in the legend. The finO gene defines the ori in the reference and is designated as the +1 start site. The order of query genomes plotted in the circle reflects their respective clade association from 1 to 9 as indicated by the colored boxes in the legend. We used the pO157 plasmids of strains EDL933 and WS4202 as reference, the latter is 636 bp larger (Latif et al., 2014). Major differences are associated with MGEs, highlighted in gray. GC-content and -skew of the pO157 EC4115 reference plasmid are depicted in the two innermost circles, respectively. (B) SNP-derived phylogeny of three virulence plasmid types. The tree was recovered using a heuristic search in PAUP (Wilgenbusch and Swofford, 2003) with 500 bootstrap replicates, and visualized in Geneious (Kearse et al., 2012). The vertical gray bar indicates the plasmid variant and the colored boxes reflect the clade as shown in the legend.