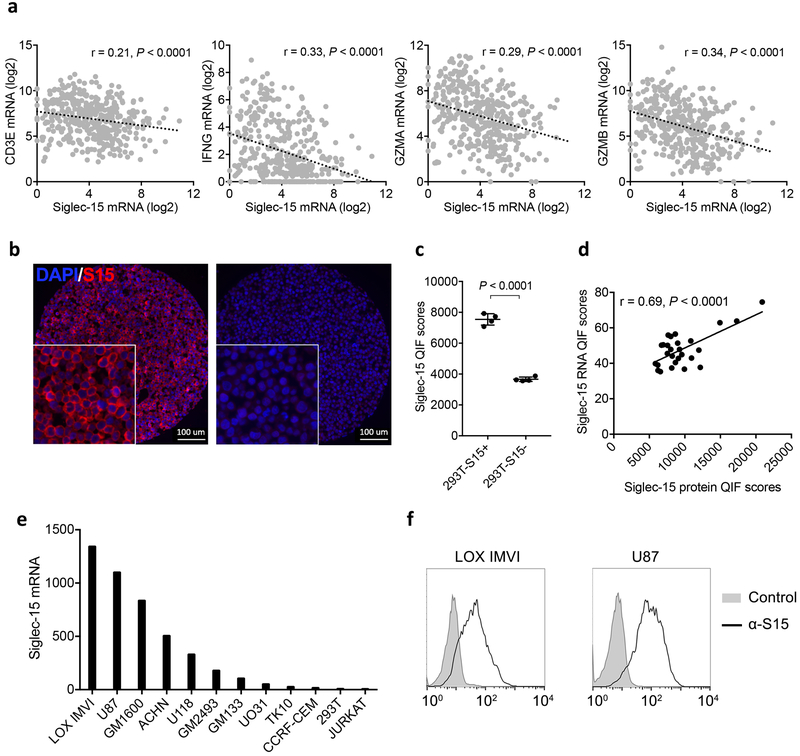
Extended Data Figure 6. Analysis of Siglec-15 mRNA expression in human cancers.
(a) The inverse correlation of mRNA expression levels between Siglec-15 and T-cell signature genes (CD3E, IFNG, GZMA and GZMB) in bladder cancer by meta-analysis of TCGA databases. Pearson r score and P value are shown (n=407 human samples).
(b-d) Validation of anti-Siglec-15 antibody clone PA5-48221. Representative quantitative immunofluorescence images of positive staining on 293T cells overexpressing Siglec-15 (293T-S15+, left panel) compared to mock transfected 293T cells (293T-S15-, right panel) (DAPI [blue] and S15 [red]) (b) Data are representative of four independent experiments. QIF scores of 293T-S15+ and 293T-S15- cell lines (c). Data are mean ± s.e.m. (n= 4 independent experiments). P values by two-tailed unpaired t-test. Comparison of Siglec-15 protein and RNA expression using RNAscope in situ detection by QIF (d). Pearson r score and P value are shown (n = 27 human samples).
(e) The relative levels of Siglec-15 mRNA in human cancer cell lines from the BioGPS database.
(f) Cell surface expression of Siglec-15 on LOX IMVI and U87 human cancer lines by staining with anti-Siglec-15 and control mAb and analyzed with flow cytometry. Data are representative of three independent experiments.