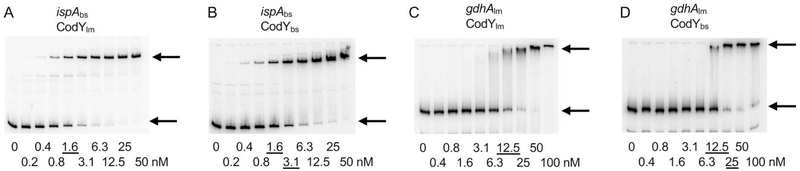
Fig. 7.
Comparison of binding by CodY proteins from L. monocytogenes and B. subtilis by gel shift assays. Radioactively labeled DNA fragments of the B. subtilis ispA gene (A, B) or the L. monocytogenes gdhA gene (C, D) were incubated with increasing amounts of purified L. monocytogenes (A, C) or B. subtilis (B, D) CodY in the presence of 10 mM ILV. CodY monomer concentrations used (nM) are indicated below each lane, and the concentrations needed to shift ~50% of DNA fragments, are underlined. The arrows indicate the bands corresponding to unbound DNA fragments (lower bands) and the complexes of CodY with DNA (upper bands).