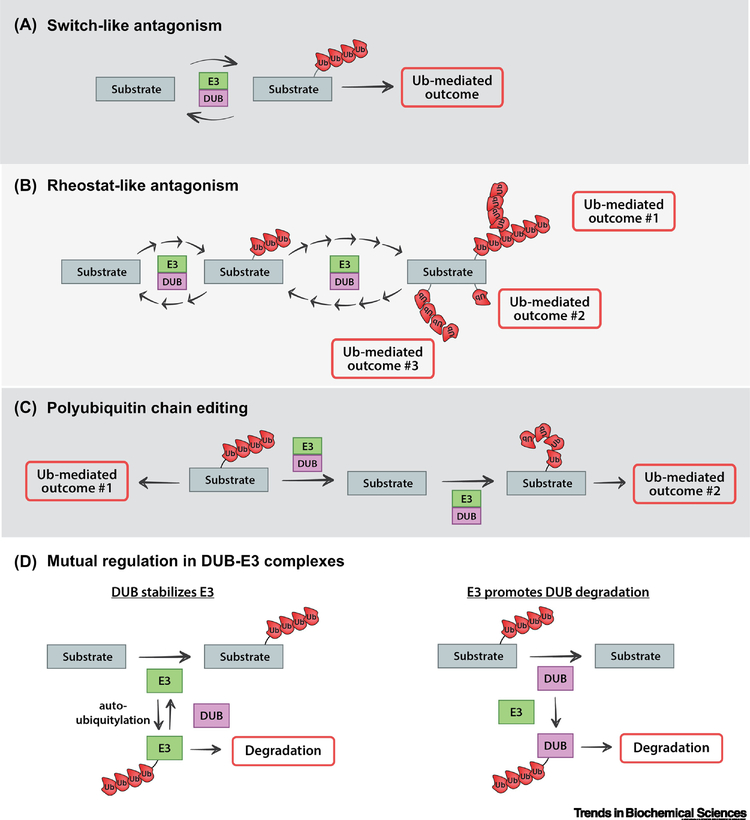
Figure 3.
The regulatory logic of coupling ubiquitin conjugation and deconjugation activities. DUB-E3 complexes can operate on a shared substrate, resulting regulation of substrate protein fate that can be switch-like or rheostat-like. (A) Switch-like behavior results from direct antagonism in a DUB-E3 complex and can lead to ubiquitin-driven outcomes that are binary in nature. In such cases, E3 activity must overcome the counter-acting DUB activity to achieve a ubiquitin-mediated outcome. (B) Rheostat-like behavior can also result from antagonism in a DUB-E3 complex when fine-tuning of E3 and DUB activities results the substrate transiting through different states with varying degrees of ubiquitylation, leading to graded responses and/or multi-variate outcomes. (C) DUB-E3 complexes can mediate polyubiquitin chain editing. This occurs when the DUB-E3 complex operates sequentially on a polyubiquitylated substrate, with the DUB activity removing the extant polyubiquitin chain and the E3 activity adding a polyubiquitin chain of a different linkage type. This type of chain remodeling can have the effect of altering the fate of the substrate protein. (D) DUB-E3 complexes can also lead to mutual regulation of DUB and E3 stability. Specifically, DUBs can protect an interacting E3 from the potential degradation that can result from autoubiquitylation (left) while an E3 can potentially ubiquitylate an interacting DUB and thereby alter its activity or promote its degradation (right).