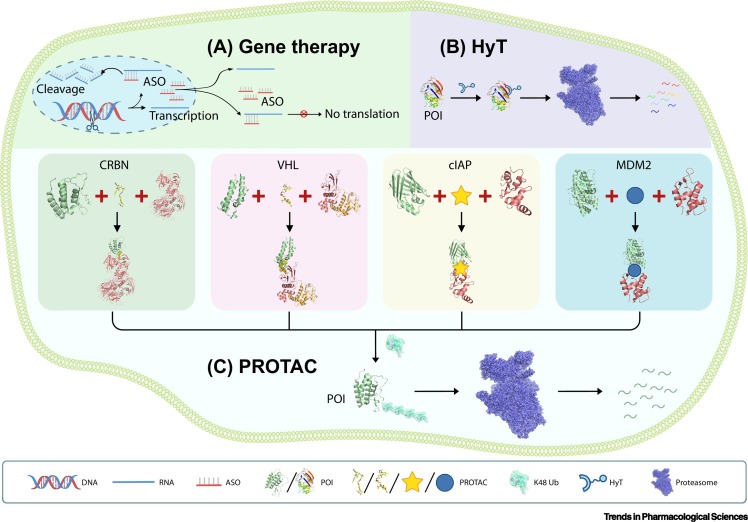
Figure 1.
Schematic Models of Established Strategies to Selectively Target a Protein of Interest (POI).
(A) Schematic illustration of DNA- and RNA-targeting technologies such as genome editing (represented by a pair of scissors cutting the target DNA) or antisense oligonucleotides (ASOs) that induce the target RNA degradation and protein translation inhibition. (B) Schematic illustration of the hydrophobic tagging (HyT) technology, which adds a hydrophobic tag onto POIs to induce their degradation via the proteasome, independently of E3 ligases and ubiquitination. (C) Schematic illustration of the four major PROTAC systems utilizing the indicated E3 ligase subunits. The PROTAC molecules bring the POIs in proximity to the corresponding E3 ligases to facilitate POI K48 polyubiquitination (Ub), leading to their subsequent degradation via the proteasome. All molecules were drawn based on public PDB files: CRBN-PROTAC-BRD4 (PDB: 6BN7); VHL-PROTAC-BRD4 (PDB: 5T35); cIAP1 (PDB: 3UW4), CRABP2 (PDB: 3CBS); MDM2 (PDB: 4HF2), AR (PDB: 1XOW); Ub (PDB: 5GOI); HyT POI DHFR (PDB: 5UII); proteasome (PDB: 4CR2). The PDB files of the cIAP and MDM2 PROTAC complexes were unavailable, and their PROTAC molecules therefore are represented by cartoon shapes.