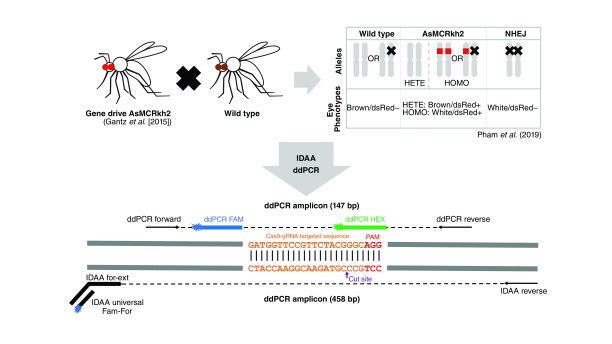
Figure 1. . Assessment of nonhomologous end joining alleles from AsMCRkh2 mosquitoes using ddPCR and IDAA techniques.
AsMCRkh2 is a gene drive transgenic Anopheles stephensi mosquito line that contains an autonomous Cas9-gRNA system linked to a dominant DsRed eye marker that targets the kynurenine hydroxylase (kh) locus [5]. WT mosquitoes and heterozygous AsMCRkh2 mosquitoes with one intact kh allele have a black-eyed/DsRed-positive phenotype, whereas homozygous AsMCRkh2 individuals have a recessive white-eyed/DsRed-positive phenotype. In contrast, mosquitoes presenting both resistant alleles present a white-eyed/DsRed-negative phenotype. NHEJ mosquitoes used in this report came from cage trials established from an outcross of AsMCRkh2 mosquitoes with WT where a susceptible kh allele was cleaved by gRNA-guided Cas9 nuclease and was repaired with NHEJ instead of HDR [5,11] to become a Cas9-resistant kh allele with mutations around the cut site. NHEJ mosquitoes from different cage generations were used for DNA extraction and PCR using primers designed to amplify a 458-bp PCR fragment and 147-bp PRC fragment spanning the targeted sequence for IDAA and ddPCR analysis, respectively.
ddPCR: Droplet Digital PCR; HDR: Homology-directed repair; IDAA: Indel detection by amplicon analysis; NHEJ: Nonhomologous end joining; WT: Wild-type.