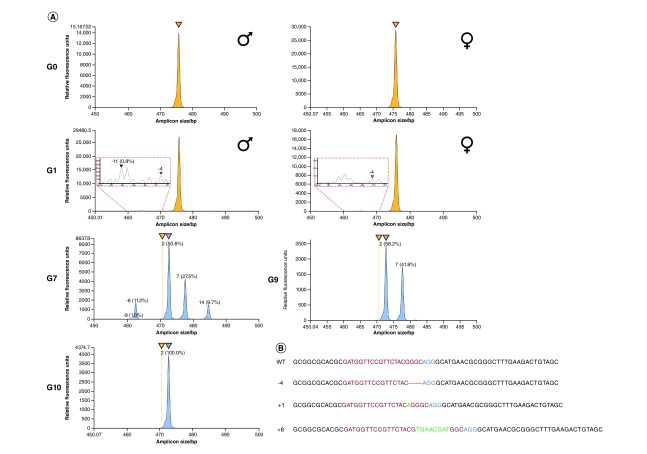
Figure 3. . Tracing and quantification of insertions or deletions in AsMCRkh2 mosquito samples over generations.
G0 and G1 mosquito samples were chosen randomly, and samples from G7 through G10 were all individuals carrying NHEJ alleles selected by phenotype (white-eyed/DsRed-negative). (A) G0: ‘Founder’ individuals show the baseline for the IDAA profile. Both males and females present a WT sequence at the target site shown by a yellow peak (because all female G0s are WT, whereas even though G0 males were a combination of transgenic and WT males only WT [and NHEJ alleles, if there were any] were amplified by PCR). G1: First-generation offspring display expected low frequency of indels in sample pools of WT and low-frequency NHEJ individuals. Red-dotted line zoom-in inserts display the rarely occurring NHEJ indel events in the population (<0.8%), and the black and gray triangles indicate spectra peaks of indels. Two types of indels, -11 and -4, were identified in G1. IDAA and ddPCR allowed the analysis of a large number of samples from G0 and G1, which was required to determine NHEJ allele-generated frequency. G7–G10: White-eyed phenotype mosquitoes with homozygous NHEJ alleles were selected from different generations. Although phenotypically similar, the variable peak heights indicate that G7 individuals represent a heterogeneous population, with different types of indels, whereas G9 (near equal peak heights) and G10 (single peak) represent a homogeneous population with only one indel (-4) selectively carried to subsequence generations. WT alleles are distinguished by yellow peaks when present in the spectra; when absent, yellow triangles above the spectra panels are used to reference the WT location. Frameshift-causing indels are indicated with peaks color-coded in blue. (B) Sanger sequencing in mosquitoes from G7 (11 individuals), G9 (5 individuals) and G10 (16 individuals) show results comparable with the IDAA findings. Three types of indels (-4, +1, +8) were identified in G7 mosquitoes, whereas only one type of indel (-4) was present in G9 and G10 (Supplementary Table 3).
ddPCR: Droplet Digital PCR; IDAA: Indel detection by amplicon analysis; Indel: Insertions or deletion; NHEJ: Nonhomologous end joining; WT: Wild-type.