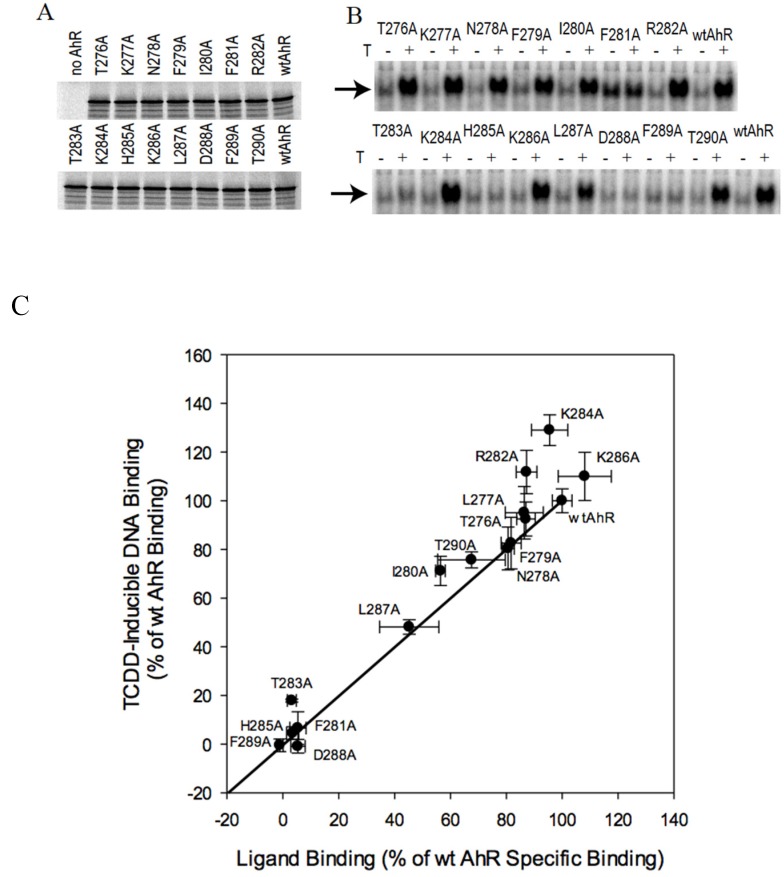
Figure 2.
Effects of mutagenesis of amino acids 276–290 of the AhR PASB on 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible AhR transformation/DNA binding and [3H]TCDD specific binding. (A) In vitro protein expression of AhR mutants. Indicated AhR constructs were synthesized in vitro in the presence of [35S]-methionine and resolved by SDS-PAGE and autoradiography. (B) In vitro synthesized wild type (wt) or mutant AhRs and ARNT were diluted at 1:1:8 (v/v/v) ratio (AhR:ARNT:buffer), transformed in the presence of 10 nM TCDD (T) or 1% DMSO (v/v) for 3 h, and protein-DNA complexes analyzed by gel retardation analysis. A representative gel is shown and the specific protein-DNA complex is indicated with an arrow. (C) Comparison of ligand binding and ligand-dependent DNA binding for wt and mutant AhRs. [3H]TCDD binding to the in vitro synthesized AhR in the presence ARNT, diluted at 1:1:8 (v/v/v) ratio (AhR:ARNT:buffer), was analyzed by hydroxyapatite assay following 1 h of incubation at room temperature in the presence of 10 nM [3H]TCDD. Values were normalized to those obtained with unprogrammed lysate (non-specific binding control) and plotted vs corresponding values of the TCDD-inducible protein-DNA complex for each AhR from gel retardation experiments (panel A). Values represent the mean ± standard deviation of triplicate independent binding reactions. Data points overlapping with the trend line would indicate a proportional change in ligand binding and DNA binding for the indicated mutant AhR (i.e., unchanged transformation efficiency; see text). Note: F281A was constitutively active in DNA binding (2B); however, since it demonstrated no ligand-dependent increase in DNA binding, the resulting ligand-dependent change in DNA binding was low and similar to that of inactive AhRs (2B). Results presented are representative of at least three independent experiments.