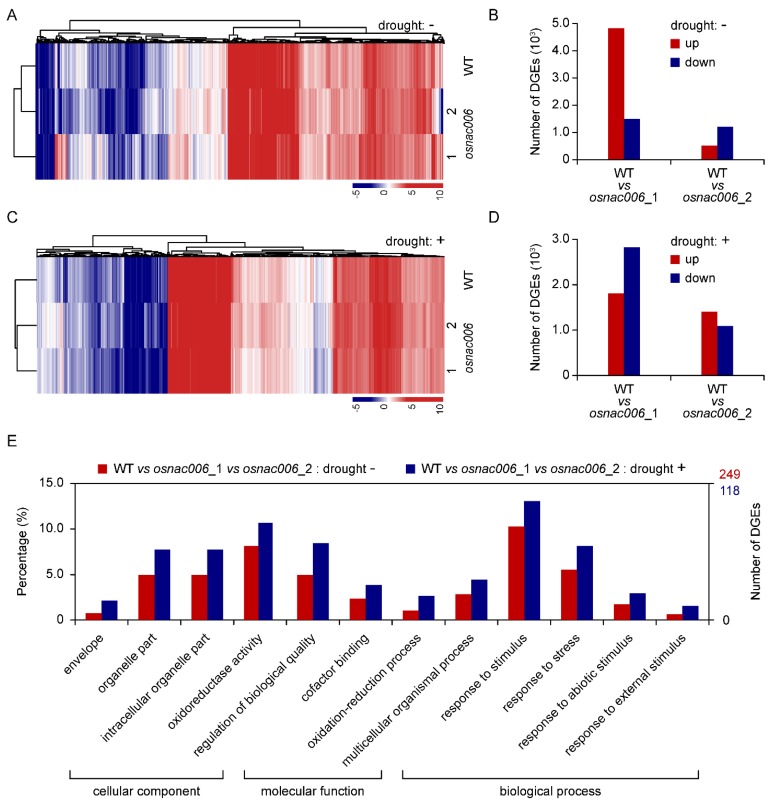
Figure 4.
Global gene expression changes in knockout OsNAC006 rice. (A) The most significant clustering analysis of differentially expressed genes (DEGs) in WT and osnac006 T1 mutant lines. Targeted knockout of osnac006 resulted in profound changes to gene expression, physiology, and development compared with WT and controls without drought stress treatment. The colour scale corresponds to log2 (FPKM) values of the genes. (B) Number of DEGs in WT, osnac006_1 and osnac006_2 T1 mutant lines, based on expression profiles obtained by RNA-Seq. Total RNA was extracted from mixed samples from three separate plants. (C) DEGs shared by WT and osnac006_1 and WT and osnac006_2 lines before drought stress. (D) DEGs shared by WT and osnac006_1 and WT and osnac006_2 lines after drought stress. (E) Gene ontology (GO) classification of DEGs shared by WT and osnac006_1 and WT and osnac006_2 mutant lines under normal and drought stress conditions. The x-axis shows user-selected GO terms, and the y-axis shows the percentage of genes (number of a particular gene divided by the number of total genes).