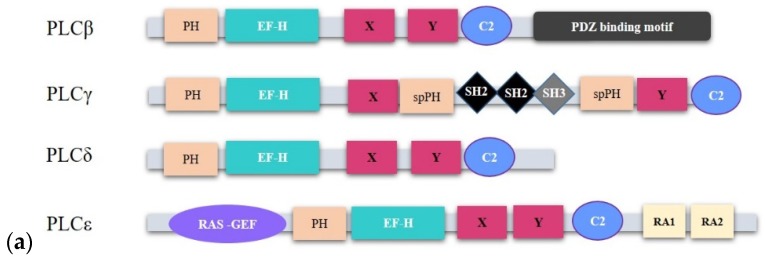
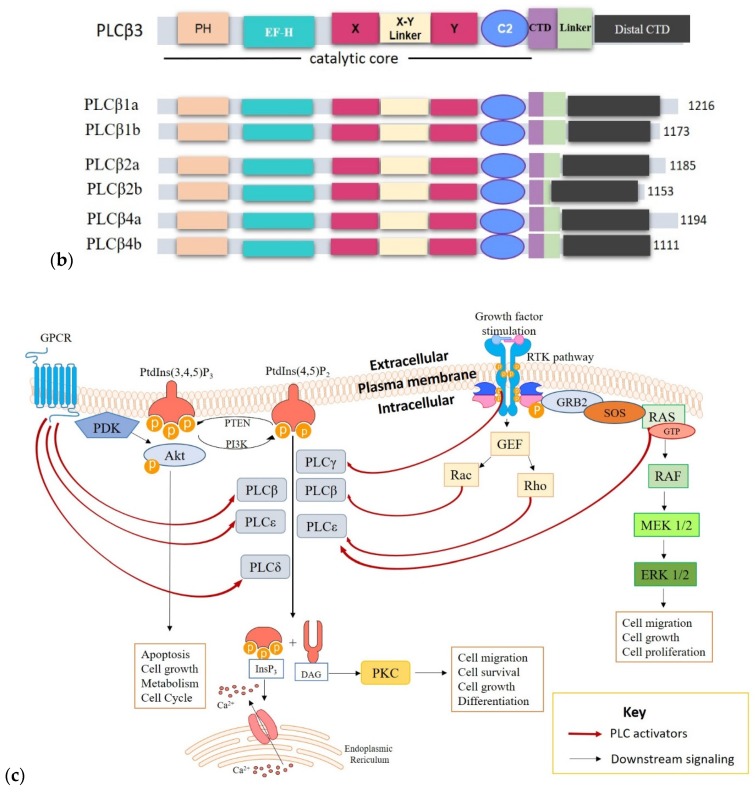
Figure 2.
The structure and activation of phospholipase C (PLC) isozymes. (a): cartoon representing the structural components of PLC family members implicated in cancer. PLC isoforms possess structurally conserved domains such as the EF, X and Y catalytic core, and the C2 domains. However, the organization of their regulatory domains follows a subtype dependent manner. (b): PLCβ isoforms and their spliced variants show conserved structural features with minor differences at the C-terminal domain (CTD). They incorporate a core set of domains consisting of an N-terminal PH domain, four EF-hand motifs, a X–Y catalytic site, a C2 domain, and a C-terminal domain with a linker between the proximal and distal ends. The various isoforms possess varied lengths and sequences occurring within the C-terminal extensions. (c): PLCs are activated by different stimuli to mediate the hydrolysis of PtdIns(4,5)P2 into the second messengers InsP3 and DAG, which subsequently promote the intracellular release of Ca2+ from the endoplasmic reticulum (ER) and activation of PKC, respectively. PKC activation following DAG and Ca2+ release promotes cell migration, cell survival and differentiation. Some PLCs are activated by more than one mechanism. For example, PLCε can be activated by the rat sarcoma (RAS) protein and the RAS homolog family member (Rho) as well as G-proteins. PLCβ, especially PLCβ2 can be activated via the classical G-protein pathway but also through RAS-related C3 botulinum toxin substrate (Rac) GTPases. However, the routes of activation may be cell type-specific or dependent on stimuli. Red-colored arrows represent reactions that activate PLCs while black arrows show downstream signaling paths.