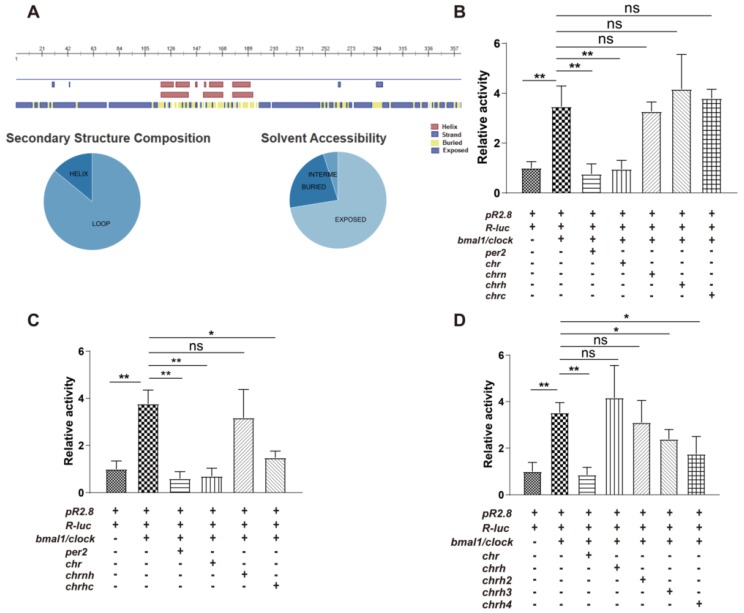
Figure 1.
CHR repression domain that contains an α-helix rich region. (A) Predicted secondary structures show an α-helix rich region in the middle of CHR, with flexible regions in the N- and C- termini. Pie charts show that CHR containing a large proportion of flexible composition and a lot of regions are exposed. (B) Based on predicted secondary structures, CHR was divided into 3 separate parts: CHRN (1–110), CHRH (111–196), and CHRC (197–385). In the classical luciferase reporter assay which uses B/C complexes to activate an E-box containing promoter (pR2.8 in this experiment), each individual part of CHR lost the repressive activity of CHR. (C) However, the combination of the predicted helix region plus the C-terminus (CHRHC, 111-385) is able to function as a repressor, rather than the combination of the helix region plus the N-terminus (CHRNH, 1-196). (D) Luciferase assays indicate that CHRH plus extended regions at its C-terminus (CHRH3, 111–264 and CHRH4, 111–298) could repress the transcription activation by B/C complexes. Since CHRH3 is 2 amino acids longer than CHRH2 that did not perform the repressive function, this indicates that the helix containing H3 is the minimal domain that functions as a repressor to B/C complexes. At least three independent experimental repeats were done for each luciferase reporter assay. Graphpad Prism 5 was used to generate graphs/plots and perform statistical analysis (2-tailed unpaired t-test). ns, no significance; * p < 0.05; ** p < 0.01.