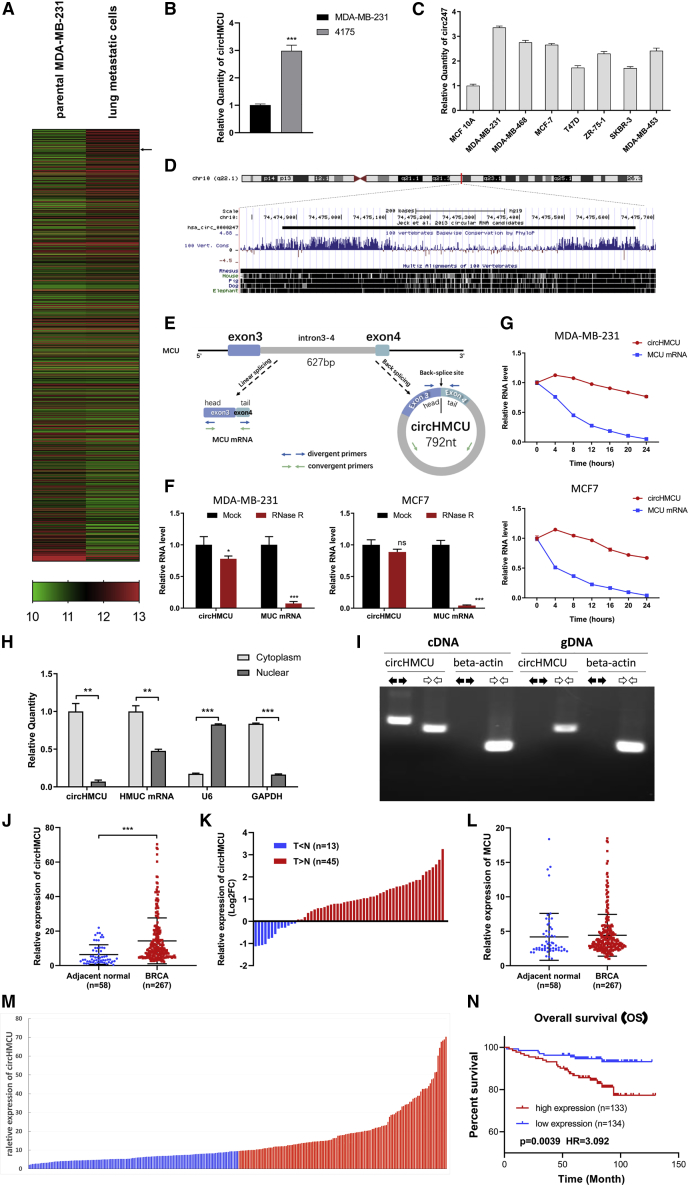
Figure 1.
Identification and Characteristics of circHMCU (hsa_circ_0000247) in Breast Cancer
(A) The cluster heatmap shows the differentially expressed circRNAs between parental MDA-MB-231 and lung metastatic cells. Red indicates a high expression level, and green indicates a low expression level. The arrow indicates circHMCU. (B) Expression level of circHMCU in MDA-MB-231 and its derivative (4175). (C) circHMCU expression in normal breast epithelial cells and breast cancer cell lines. (D) circHMCU is derived from the third and fourth exons as well as the intron between them from the human MCU gene, which is located on chromosome 10. (E) Scheme illustrating the production of circHMCU. Arrow represents “head-to-tail” splicing sites of circHMCU. (F) Quantitative real-time PCR analysis of circHMCU and MCU mRNA after RNase R treatment. (G) Quantitative real-time PCR analysis of circHMCU and MCU mRNA after ActD treatment. DMSO treatment was performed as a negative control. (H) Quantitative real-time PCR analysis of circHMCU and MCU mRNA in the cytoplasm or the nucleus in MCF-7 cells. (I) The existence of circHMCU was validated in breast cancer cells by PCR. Divergent primers amplified circHMCU in cDNA but not genomic DNA (gDNA). β-Actin was used as negative control. (J) The expression level of circHMCU was increased in breast cancer tissues (n = 267) compared with normal breast tissues (n = 58). (K) 77.59% (43/58) breast cancer patients presented increased expression of circHMCU. The bar chart presents the log2FC (fold change). (L) Expression level of MCU (linear isoform) in breast cancer tissues (n = 267) compared with normal breast tissues (n = 58). (M) According to the median expression level of circHMCU, the 267 breast cancer patients were divided into two groups. The blue bars represent the low-expression group, and the red bars represent the high-expression group. (N) Kaplan-Meier analysis of the association between circHMCU expression and prognosis of breast cancer patients. Data are the means ± SEM of three experiments. ∗∗p < 0.01, ∗∗∗p < 0.001.